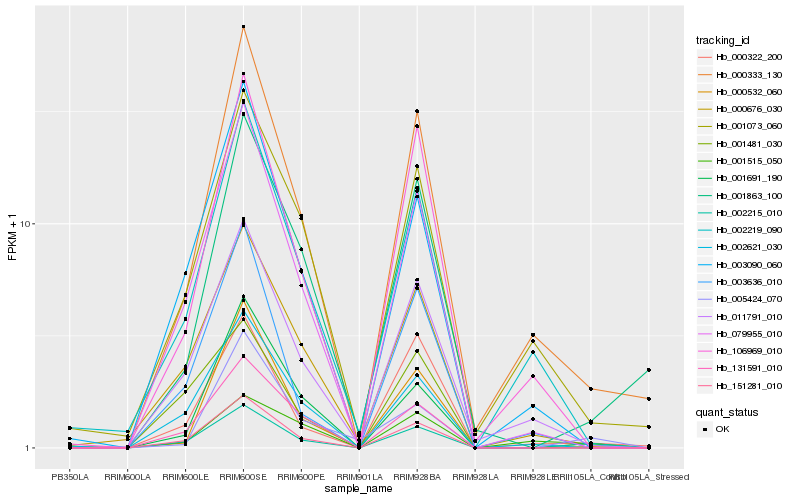
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002215_010 |
0.0 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 2 |
Hb_151281_010 |
0.1573918241 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable L-type lectin-domain containing receptor kinase S.5 [Jatropha curcas] |
| 3 |
Hb_001481_030 |
0.158504909 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 4 |
Hb_079955_010 |
0.1644395392 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-related protein Myb4-like [Populus euphratica] |
| 5 |
Hb_000333_130 |
0.1752241177 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 6 |
Hb_131591_010 |
0.1753377409 |
- |
- |
PREDICTED: cytochrome P450 83B1-like [Jatropha curcas] |
| 7 |
Hb_000676_030 |
0.1761882085 |
- |
- |
PREDICTED: probable mitochondrial chaperone BCS1-B [Jatropha curcas] |
| 8 |
Hb_002621_030 |
0.181311303 |
- |
- |
hypothetical protein CISIN_1g006155mg [Citrus sinensis] |
| 9 |
Hb_011791_010 |
0.1840004676 |
- |
- |
PREDICTED: cytochrome P450 83B1-like [Jatropha curcas] |
| 10 |
Hb_003090_060 |
0.1860264912 |
- |
- |
PAR1 protein [Theobroma cacao] |
| 11 |
Hb_000322_200 |
0.1870615034 |
transcription factor |
TF Family: bHLH |
PREDICTED: putative transcription factor bHLH041 [Jatropha curcas] |
| 12 |
Hb_001863_100 |
0.1985793173 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_005424_070 |
0.1994510278 |
- |
- |
PREDICTED: TATA-box-binding protein 1-like [Jatropha curcas] |
| 14 |
Hb_001073_060 |
0.2010678727 |
- |
- |
phosphate transporter 1 [Hevea brasiliensis] |
| 15 |
Hb_000532_060 |
0.2045609705 |
- |
- |
copper-transporting atpase p-type, putative [Ricinus communis] |
| 16 |
Hb_003636_010 |
0.2060783842 |
- |
- |
hypothetical protein CISIN_1g037850mg, partial [Citrus sinensis] |
| 17 |
Hb_106969_010 |
0.2062193905 |
- |
- |
Cytochrome P450 76C4 [Glycine soja] |
| 18 |
Hb_001691_190 |
0.2078016651 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002219_090 |
0.2084783403 |
- |
- |
protein kinase atmrk1, putative [Ricinus communis] |
| 20 |
Hb_001515_050 |
0.209494679 |
- |
- |
PREDICTED: uncharacterized protein LOC105636626 [Jatropha curcas] |