| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
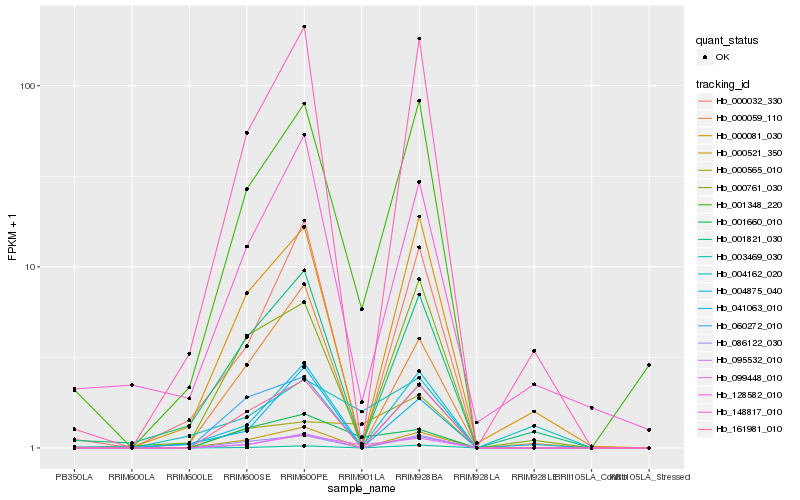
Hb_099448_010 |
0.0 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 2 |
Hb_001660_010 |
0.1566561028 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001348_220 |
0.1810659258 |
- |
- |
PREDICTED: defensin Ec-AMP-D2-like [Nelumbo nucifera] |
| 4 |
Hb_095532_010 |
0.233169422 |
- |
- |
BnaC05g27570D [Brassica napus] |
| 5 |
Hb_000565_010 |
0.2353022593 |
- |
- |
hypothetical protein JCGZ_03337 [Jatropha curcas] |
| 6 |
Hb_004875_040 |
0.237303171 |
- |
- |
PREDICTED: uncharacterized protein LOC105645797 [Jatropha curcas] |
| 7 |
Hb_161981_010 |
0.2388934101 |
- |
- |
brassinosteroid insensitive 1 -like protein [Gossypium arboreum] |
| 8 |
Hb_000521_350 |
0.2399297152 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 9 |
Hb_148817_010 |
0.2400883934 |
- |
- |
PREDICTED: CASP-like protein 1D1 [Jatropha curcas] |
| 10 |
Hb_086122_030 |
0.2464505358 |
- |
- |
D-glycerate 3-kinase, chloroplast precursor, putative [Ricinus communis] |
| 11 |
Hb_000032_330 |
0.2508138595 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_128582_010 |
0.2566155913 |
- |
- |
- |
| 13 |
Hb_004162_020 |
0.2571163627 |
- |
- |
- |
| 14 |
Hb_000059_110 |
0.2593722559 |
transcription factor |
TF Family: MYB |
MYB transcriptional factor [Populus tremula x Populus tremuloides] |
| 15 |
Hb_001821_030 |
0.2594129668 |
- |
- |
hypothetical protein JCGZ_13357 [Jatropha curcas] |
| 16 |
Hb_000081_030 |
0.2607972935 |
- |
- |
BnaC09g05810D [Brassica napus] |
| 17 |
Hb_000761_030 |
0.2613746013 |
transcription factor |
TF Family: ERF |
Transcriptional factor TINY, putative [Ricinus communis] |
| 18 |
Hb_060272_010 |
0.262623641 |
- |
- |
PREDICTED: gibberellin 20 oxidase 2 [Populus euphratica] |
| 19 |
Hb_041063_010 |
0.2636257499 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 20 |
Hb_003469_030 |
0.2651337902 |
- |
- |
hypothetical protein JCGZ_01307 [Jatropha curcas] |