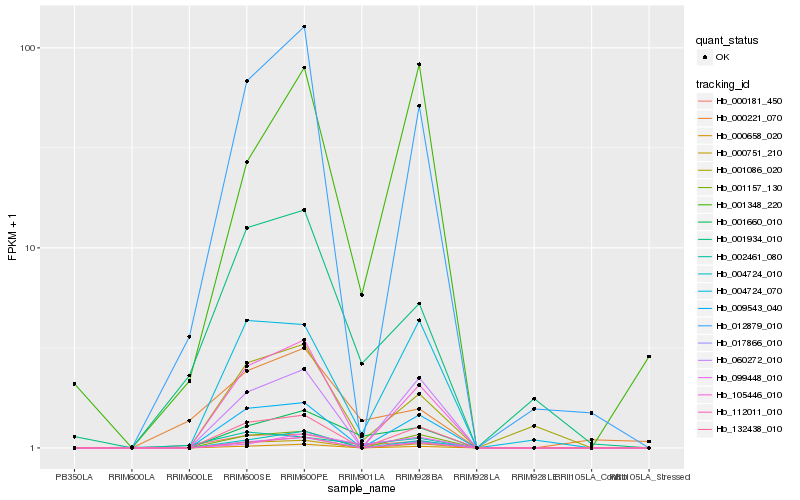
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001660_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_099448_010 |
0.1566561028 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 3 |
Hb_000181_450 |
0.2136453648 |
- |
- |
hypothetical protein JCGZ_20984 [Jatropha curcas] |
| 4 |
Hb_004724_070 |
0.2426313673 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor TGA2 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001934_010 |
0.2449551774 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 6 |
Hb_112011_010 |
0.2476924177 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_017866_010 |
0.2482954325 |
- |
- |
kinase family protein [Populus trichocarpa] |
| 8 |
Hb_000658_020 |
0.2494342545 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 1 [Musa acuminata subsp. malaccensis] |
| 9 |
Hb_004724_010 |
0.24986302 |
- |
- |
- |
| 10 |
Hb_002461_080 |
0.2517971159 |
- |
- |
PREDICTED: probable pectinesterase 53 [Jatropha curcas] |
| 11 |
Hb_105446_010 |
0.251814087 |
- |
- |
- |
| 12 |
Hb_000751_210 |
0.2538747417 |
- |
- |
PREDICTED: petal death protein isoform X2 [Jatropha curcas] |
| 13 |
Hb_132438_010 |
0.2547486893 |
- |
- |
PREDICTED: uncharacterized protein LOC104800023, partial [Tarenaya hassleriana] |
| 14 |
Hb_001086_020 |
0.2586866069 |
- |
- |
hypothetical protein POPTR_0013s01730g [Populus trichocarpa] |
| 15 |
Hb_001157_130 |
0.2613429301 |
- |
- |
PREDICTED: transmembrane protein 45B-like [Jatropha curcas] |
| 16 |
Hb_009543_040 |
0.2614430605 |
- |
- |
- |
| 17 |
Hb_001348_220 |
0.2614645505 |
- |
- |
PREDICTED: defensin Ec-AMP-D2-like [Nelumbo nucifera] |
| 18 |
Hb_012879_010 |
0.2633235434 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_060272_010 |
0.2644436862 |
- |
- |
PREDICTED: gibberellin 20 oxidase 2 [Populus euphratica] |
| 20 |
Hb_000221_070 |
0.2668823708 |
- |
- |
conserved hypothetical protein [Ricinus communis] |