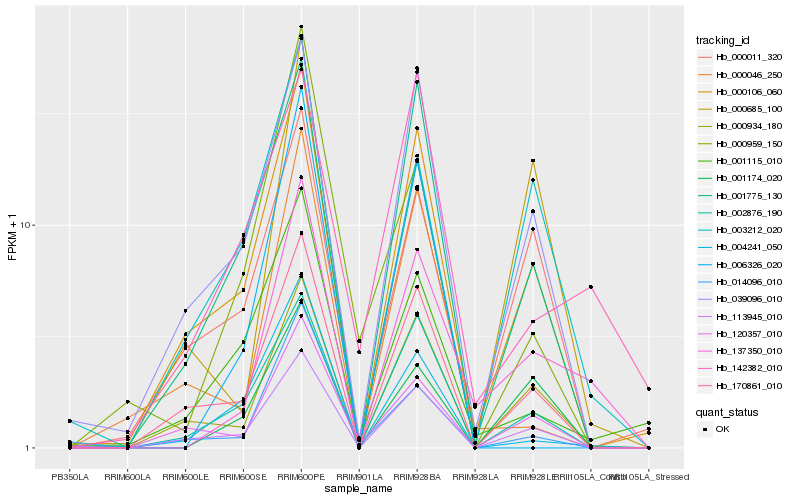
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_137350_010 |
0.0 |
- |
- |
PREDICTED: potassium channel SKOR-like [Prunus mume] |
| 2 |
Hb_000106_060 |
0.2120774504 |
- |
- |
PREDICTED: uncharacterized protein LOC105636580 [Jatropha curcas] |
| 3 |
Hb_000934_180 |
0.2202238826 |
- |
- |
hypothetical protein JCGZ_25237 [Jatropha curcas] |
| 4 |
Hb_170861_010 |
0.2280842934 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 5 |
Hb_113945_010 |
0.2312354152 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 6 |
Hb_039096_010 |
0.2343610716 |
- |
- |
unknown [Populus trichocarpa] |
| 7 |
Hb_003212_020 |
0.235641991 |
- |
- |
hypothetical protein PHAVU_009G114800g [Phaseolus vulgaris] |
| 8 |
Hb_001174_020 |
0.2375989219 |
- |
- |
- |
| 9 |
Hb_001775_130 |
0.2392399968 |
- |
- |
Prephenate dehydratase [Hevea brasiliensis] |
| 10 |
Hb_120357_010 |
0.2431128125 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 11 |
Hb_002876_190 |
0.2451870474 |
transcription factor |
TF Family: GRAS |
hypothetical protein PRUPE_ppa017519mg [Prunus persica] |
| 12 |
Hb_000685_100 |
0.2461802871 |
- |
- |
hypothetical protein POPTR_0001s10630g [Populus trichocarpa] |
| 13 |
Hb_001115_010 |
0.247231615 |
- |
- |
PREDICTED: 4-coumarate--CoA ligase 2-like [Jatropha curcas] |
| 14 |
Hb_000046_250 |
0.2498812482 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000011_320 |
0.2500872236 |
- |
- |
PREDICTED: uncharacterized protein LOC105631328 [Jatropha curcas] |
| 16 |
Hb_142382_010 |
0.2520920699 |
rubber biosynthesis |
Gene Name: Hydroxymethylglutaryl coenzyme A reductase |
hydroxymethylglutaryl-CoA reductase [Hevea brasiliensis] |
| 17 |
Hb_014096_010 |
0.2532245242 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 18 |
Hb_004241_050 |
0.2532671541 |
- |
- |
Indole-3-acetic acid-amido synthetase GH3.6, putative [Ricinus communis] |
| 19 |
Hb_006326_020 |
0.2560834144 |
- |
- |
- |
| 20 |
Hb_000959_150 |
0.2592663628 |
- |
- |
PREDICTED: uncharacterized protein LOC104879927 [Vitis vinifera] |