| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
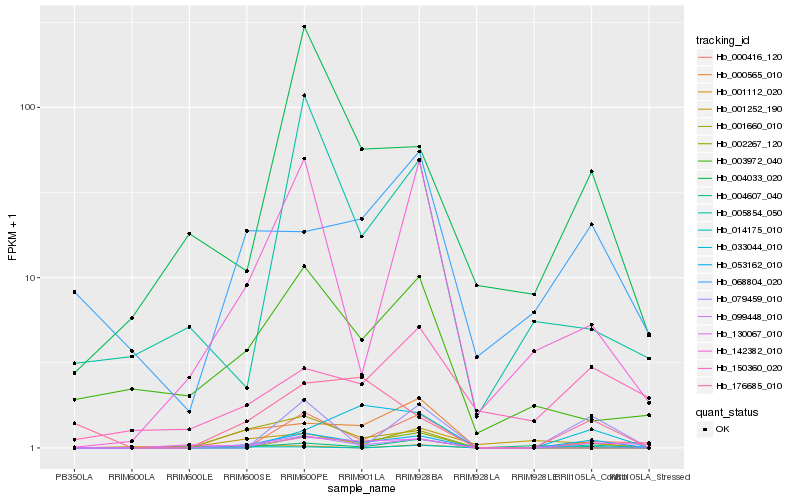
Hb_053162_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |
| 2 |
Hb_000416_120 |
0.3152564134 |
- |
- |
Putative LRR receptor-like serine/threonine-protein kinase, partial [Glycine soja] |
| 3 |
Hb_130067_010 |
0.3154927402 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 4 |
Hb_099448_010 |
0.316692291 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 5 |
Hb_079459_010 |
0.3169973507 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_142382_010 |
0.3252379897 |
rubber biosynthesis |
Gene Name: Hydroxymethylglutaryl coenzyme A reductase |
hydroxymethylglutaryl-CoA reductase [Hevea brasiliensis] |
| 7 |
Hb_004033_020 |
0.3373365581 |
transcription factor |
TF Family: MYB-related |
hypothetical protein PRUPE_ppa014728mg [Prunus persica] |
| 8 |
Hb_176685_010 |
0.3475714584 |
- |
- |
receptor-type protein kinase LRK1-like [Oryza sativa Japonica Group] |
| 9 |
Hb_033044_010 |
0.3544031733 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 5-like isoform X3 [Gossypium raimondii] |
| 10 |
Hb_004607_040 |
0.3576778326 |
- |
- |
ADP,ATP carrier protein, putative [Ricinus communis] |
| 11 |
Hb_068804_020 |
0.3579680048 |
- |
- |
hypothetical protein AMTR_s00046p00231970 [Amborella trichopoda] |
| 12 |
Hb_001660_010 |
0.3624853134 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_014175_010 |
0.3662568169 |
- |
- |
Uncharacterized protein TCM_033221 [Theobroma cacao] |
| 14 |
Hb_000565_010 |
0.3693555727 |
- |
- |
hypothetical protein JCGZ_03337 [Jatropha curcas] |
| 15 |
Hb_003972_040 |
0.3697119635 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 6 [Jatropha curcas] |
| 16 |
Hb_005854_050 |
0.3699489812 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_150360_020 |
0.3700966682 |
- |
- |
PREDICTED: N-alpha-acetyltransferase 11 [Jatropha curcas] |
| 18 |
Hb_001112_020 |
0.3719471845 |
- |
- |
PREDICTED: uncharacterized protein LOC103717724, partial [Phoenix dactylifera] |
| 19 |
Hb_001252_190 |
0.3755554992 |
desease resistance |
Gene Name: NB-ARC |
- |
| 20 |
Hb_002267_120 |
0.3769384402 |
- |
- |
hypothetical protein B456_009G136300 [Gossypium raimondii] |