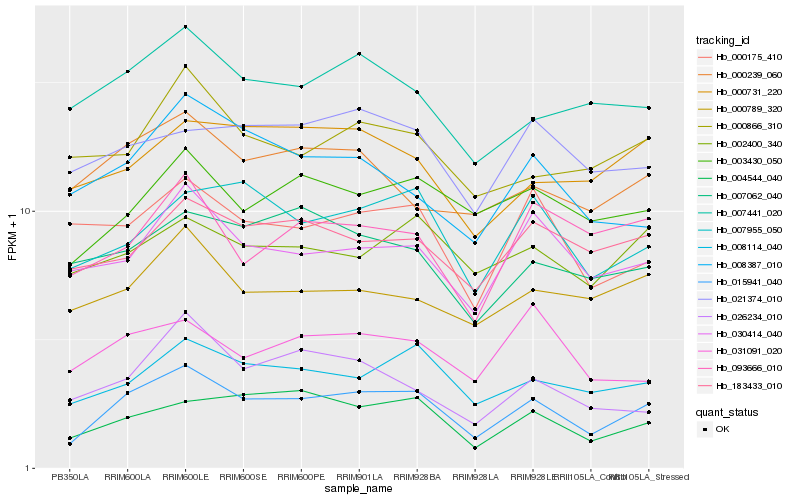
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_015941_040 |
0.0 |
- |
- |
L-galactono-1,4-lactone dehydrogenase, mitochondrial [Cucumis sativus] |
| 2 |
Hb_008114_040 |
0.1112329125 |
- |
- |
Fanconi anemia group D2 [Gossypium arboreum] |
| 3 |
Hb_004544_040 |
0.1143148917 |
- |
- |
hypothetical protein PHAVU_010G103000g [Phaseolus vulgaris] |
| 4 |
Hb_007955_050 |
0.1194108879 |
- |
- |
PREDICTED: multiple RNA-binding domain-containing protein 1 [Jatropha curcas] |
| 5 |
Hb_030414_040 |
0.1233578218 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH3 isoform X1 [Jatropha curcas] |
| 6 |
Hb_003430_050 |
0.1243470643 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor RSZ21A isoform X1 [Phoenix dactylifera] |
| 7 |
Hb_007441_020 |
0.1311857643 |
- |
- |
Rab6 [Hevea brasiliensis] |
| 8 |
Hb_093666_010 |
0.1335253195 |
- |
- |
BnaC05g06620D [Brassica napus] |
| 9 |
Hb_183433_010 |
0.1337683851 |
- |
- |
PREDICTED: FAD synthase isoform X3 [Jatropha curcas] |
| 10 |
Hb_026234_010 |
0.1343069287 |
- |
- |
choline monooxygenase, putative [Ricinus communis] |
| 11 |
Hb_008387_010 |
0.1346727878 |
- |
- |
SMAD/FHA domain-containing protein isoform 3 [Theobroma cacao] |
| 12 |
Hb_000731_220 |
0.135079485 |
- |
- |
PREDICTED: derlin-2.2 [Jatropha curcas] |
| 13 |
Hb_000239_060 |
0.1356180073 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000789_320 |
0.1363815723 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_000175_410 |
0.1365509332 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 56-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_000866_310 |
0.1370198314 |
- |
- |
PREDICTED: neural Wiskott-Aldrich syndrome protein [Jatropha curcas] |
| 17 |
Hb_002400_340 |
0.1374101286 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 10 [Jatropha curcas] |
| 18 |
Hb_021374_010 |
0.1376547954 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor NAM-A1 [Populus euphratica] |
| 19 |
Hb_031091_020 |
0.1376885406 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 20 |
Hb_007062_040 |
0.1376901 |
- |
- |
ADP-ribosylation factor GTPase-activating protein AGD2 [Morus notabilis] |