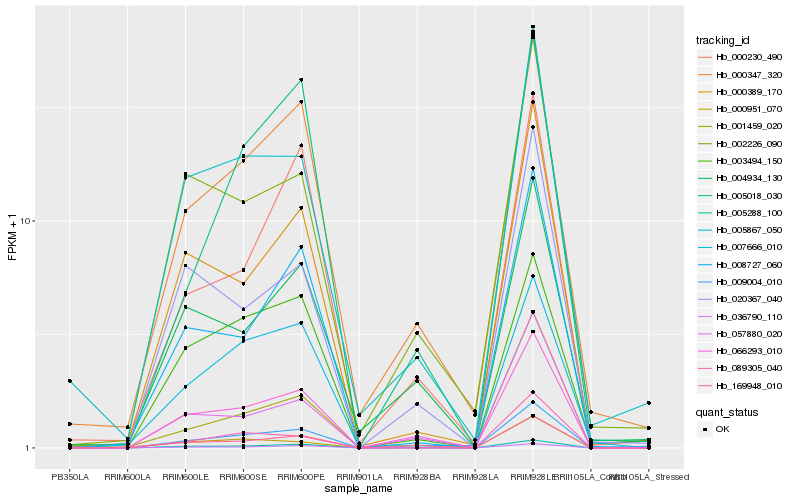
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009004_010 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At3g14460 [Jatropha curcas] |
| 2 |
Hb_005288_100 |
0.1278390011 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Prunus mume] |
| 3 |
Hb_066293_010 |
0.1278770627 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 4 |
Hb_000389_170 |
0.1304452997 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 5 |
Hb_005867_050 |
0.1334243954 |
- |
- |
PREDICTED: protein ECERIFERUM 1 [Jatropha curcas] |
| 6 |
Hb_000347_320 |
0.1356105301 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 7 |
Hb_001459_020 |
0.1404389166 |
- |
- |
ankyrin repeat protein [Bruguiera gymnorhiza] |
| 8 |
Hb_036790_110 |
0.1423489194 |
- |
- |
hypothetical protein JCGZ_22271 [Jatropha curcas] |
| 9 |
Hb_008727_060 |
0.1439021708 |
- |
- |
Beta-glucosidase, putative [Ricinus communis] |
| 10 |
Hb_003494_150 |
0.1466695831 |
- |
- |
Silicon transporter, putative [Ricinus communis] |
| 11 |
Hb_007666_010 |
0.1479792786 |
- |
- |
PREDICTED: uncharacterized protein LOC105649266 [Jatropha curcas] |
| 12 |
Hb_005018_030 |
0.1510629295 |
- |
- |
hypothetical protein CICLE_v10019811mg [Citrus clementina] |
| 13 |
Hb_000951_070 |
0.1536905203 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: receptor-like protein kinase FERONIA [Populus euphratica] |
| 14 |
Hb_057880_020 |
0.1558796732 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1, partial [Jatropha curcas] |
| 15 |
Hb_000230_490 |
0.1581518235 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002226_090 |
0.1600144399 |
- |
- |
Stachyose synthase precursor, putative [Ricinus communis] |
| 17 |
Hb_089305_040 |
0.1601287191 |
- |
- |
- |
| 18 |
Hb_169948_010 |
0.1612539399 |
- |
- |
hypothetical protein VITISV_006524 [Vitis vinifera] |
| 19 |
Hb_020367_040 |
0.1623752599 |
- |
- |
hypothetical protein JCGZ_01061 [Jatropha curcas] |
| 20 |
Hb_004934_130 |
0.1638257576 |
- |
- |
12-oxophytodienoate reductase 2 isoform 1 [Theobroma cacao] |