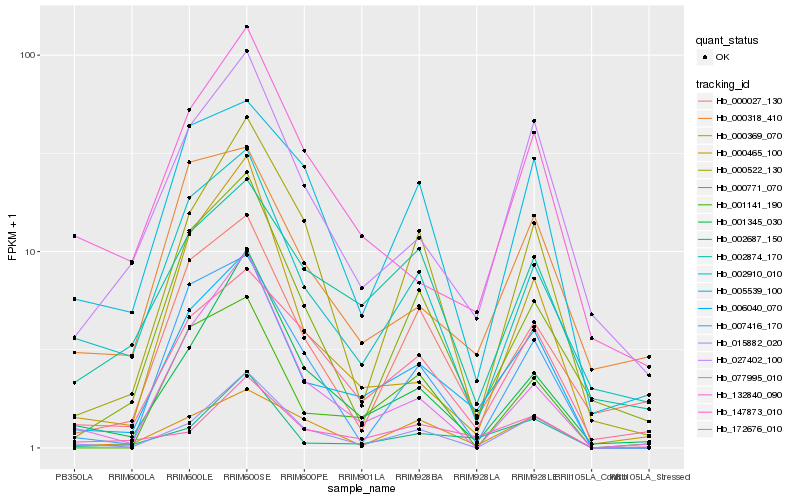
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006040_070 |
0.0 |
- |
- |
orf107b gene product (mitochondrion) [Beta vulgaris subsp. vulgaris] |
| 2 |
Hb_027402_100 |
0.1621354315 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein JCGZ_11638 [Jatropha curcas] |
| 3 |
Hb_002687_150 |
0.1649622976 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 4 |
Hb_172676_010 |
0.1692464805 |
- |
- |
PREDICTED: putative disease resistance protein At3g14460 [Jatropha curcas] |
| 5 |
Hb_002910_010 |
0.1765356713 |
- |
- |
PREDICTED: uncharacterized protein LOC105635519 [Jatropha curcas] |
| 6 |
Hb_001345_030 |
0.1775319831 |
- |
- |
hypothetical protein POPTR_0007s10390g, partial [Populus trichocarpa] |
| 7 |
Hb_015882_020 |
0.1780593664 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat containing protein, putative [Ricinus communis] |
| 8 |
Hb_000465_100 |
0.1794136762 |
- |
- |
catalytic, putative [Ricinus communis] |
| 9 |
Hb_007416_170 |
0.1864477176 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 17 [Jatropha curcas] |
| 10 |
Hb_001141_190 |
0.1870877216 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000771_070 |
0.1874582632 |
- |
- |
PREDICTED: plastidial pyruvate kinase 4, chloroplastic [Jatropha curcas] |
| 12 |
Hb_147873_010 |
0.1888590133 |
- |
- |
PREDICTED: uncharacterized protein LOC102595311 [Solanum tuberosum] |
| 13 |
Hb_000369_070 |
0.1900418285 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 14 |
Hb_000318_410 |
0.1924538138 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 1-like [Jatropha curcas] |
| 15 |
Hb_132840_090 |
0.1931199272 |
transcription factor |
TF Family: Pseudo ARR-B |
PREDICTED: two-component response regulator-like APRR3 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000522_130 |
0.1933442115 |
- |
- |
PREDICTED: uncharacterized protein LOC105632103 [Jatropha curcas] |
| 17 |
Hb_077995_010 |
0.19426901 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 18 |
Hb_002874_170 |
0.1947778117 |
- |
- |
PREDICTED: L-arabinokinase-like isoform X3 [Jatropha curcas] |
| 19 |
Hb_000027_130 |
0.195256252 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 [Jatropha curcas] |
| 20 |
Hb_005539_100 |
0.1953417513 |
- |
- |
nucleoside transporter, putative [Ricinus communis] |