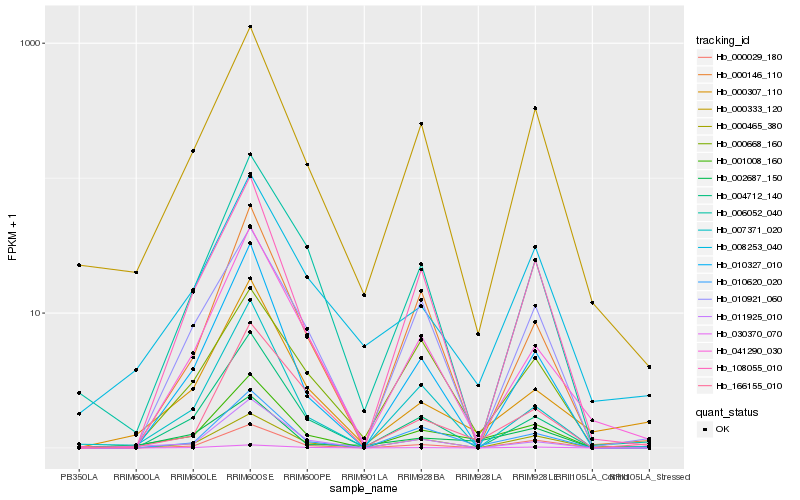
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001008_160 |
0.0 |
- |
- |
- |
| 2 |
Hb_010620_020 |
0.1573580204 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 3 |
Hb_000333_120 |
0.1626799184 |
- |
- |
PREDICTED: thiol protease aleurain-like [Jatropha curcas] |
| 4 |
Hb_002687_150 |
0.1692357857 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 5 |
Hb_000465_380 |
0.1712711313 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 6 |
Hb_000029_180 |
0.1731130034 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 7 |
Hb_011925_010 |
0.1750321152 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_108055_010 |
0.1781626559 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 9 |
Hb_010327_010 |
0.1789643448 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 10 |
Hb_030370_070 |
0.1791729078 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 11 |
Hb_004712_140 |
0.1818099954 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 12 |
Hb_041290_030 |
0.1830959549 |
- |
- |
PREDICTED: uncharacterized protein LOC105640874 [Jatropha curcas] |
| 13 |
Hb_000146_110 |
0.1855464222 |
- |
- |
resveratrol/hydroxycinnamic acid O-glucosyltransferase [Vitis labrusca] |
| 14 |
Hb_007371_020 |
0.1862180593 |
- |
- |
PREDICTED: ent-kaurenoic acid oxidase 1-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_000668_160 |
0.1875746695 |
- |
- |
PREDICTED: sulfate transporter 1.3-like [Jatropha curcas] |
| 16 |
Hb_166155_010 |
0.1878583413 |
- |
- |
PREDICTED: metalloendoproteinase 1 [Jatropha curcas] |
| 17 |
Hb_000307_110 |
0.1890638471 |
- |
- |
hypothetical protein JCGZ_08160 [Jatropha curcas] |
| 18 |
Hb_008253_040 |
0.189867999 |
- |
- |
PREDICTED: UPF0392 protein RCOM_0530710 [Jatropha curcas] |
| 19 |
Hb_006052_040 |
0.1906159172 |
- |
- |
pyruvate decarboxylase [Hevea brasiliensis] |
| 20 |
Hb_010921_060 |
0.1909839073 |
- |
- |
ATP binding protein, putative [Ricinus communis] |