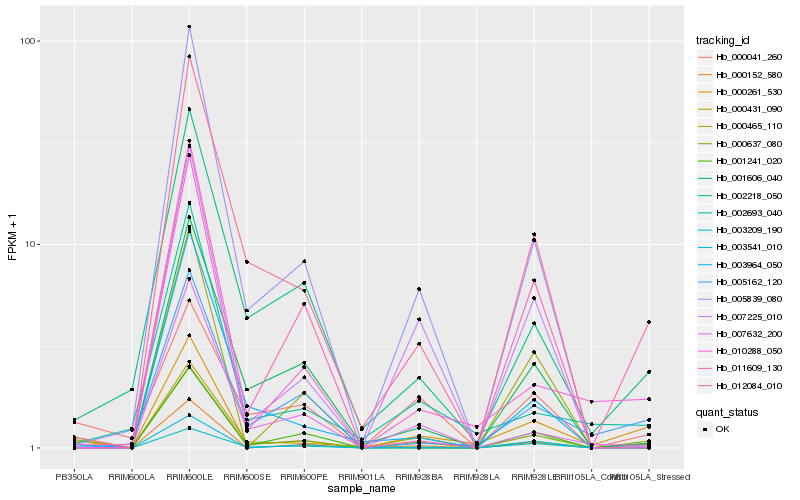
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000637_080 |
0.0 |
- |
- |
PREDICTED: protein YLS3 [Jatropha curcas] |
| 2 |
Hb_003209_190 |
0.1801782279 |
- |
- |
hypothetical protein POPTR_0010s16770g [Populus trichocarpa] |
| 3 |
Hb_000431_090 |
0.1829355407 |
- |
- |
hypothetical protein POPTR_0017s14020g [Populus trichocarpa] |
| 4 |
Hb_000041_260 |
0.2176916938 |
- |
- |
PREDICTED: reticulocalbin-2 [Jatropha curcas] |
| 5 |
Hb_002218_050 |
0.2184947523 |
- |
- |
SSXT family protein isoform 2, partial [Theobroma cacao] |
| 6 |
Hb_000261_530 |
0.2231597087 |
- |
- |
PREDICTED: beta-glucosidase 18-like [Jatropha curcas] |
| 7 |
Hb_003541_010 |
0.2264025022 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 8 |
Hb_005839_080 |
0.2298000474 |
- |
- |
Transcription factor bHLH61 isoform 1 [Theobroma cacao] |
| 9 |
Hb_011609_130 |
0.2353479876 |
- |
- |
- |
| 10 |
Hb_001241_020 |
0.2376812067 |
- |
- |
PREDICTED: uncharacterized protein LOC105628311 [Jatropha curcas] |
| 11 |
Hb_007632_200 |
0.2426205712 |
- |
- |
hypothetical protein L484_018221 [Morus notabilis] |
| 12 |
Hb_005162_120 |
0.2447106448 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |
| 13 |
Hb_010288_050 |
0.2447725979 |
- |
- |
PREDICTED: uncharacterized protein LOC105642604 [Jatropha curcas] |
| 14 |
Hb_012084_010 |
0.2451375703 |
- |
- |
cold regulated protein [Manihot esculenta] |
| 15 |
Hb_000465_110 |
0.247168012 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 330-like [Jatropha curcas] |
| 16 |
Hb_002693_040 |
0.2476743456 |
- |
- |
hypersensitive-induced response protein [Carica papaya] |
| 17 |
Hb_003964_050 |
0.2478288007 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor 25-like [Jatropha curcas] |
| 18 |
Hb_001606_040 |
0.2483589837 |
- |
- |
Rho GDP-dissociation inhibitor, putative [Ricinus communis] |
| 19 |
Hb_007225_010 |
0.2492032529 |
- |
- |
PREDICTED: glycerol-3-phosphate 2-O-acyltransferase 6 [Populus euphratica] |
| 20 |
Hb_000152_580 |
0.2499490002 |
- |
- |
PREDICTED: 21 kDa protein-like [Jatropha curcas] |