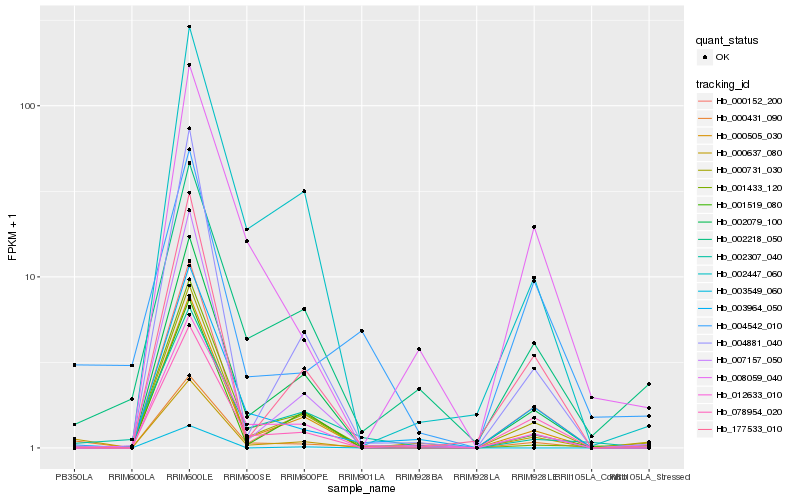
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000431_090 |
0.0 |
- |
- |
hypothetical protein POPTR_0017s14020g [Populus trichocarpa] |
| 2 |
Hb_000637_080 |
0.1829355407 |
- |
- |
PREDICTED: protein YLS3 [Jatropha curcas] |
| 3 |
Hb_003964_050 |
0.1855942359 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor 25-like [Jatropha curcas] |
| 4 |
Hb_000731_030 |
0.1976666047 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 5 |
Hb_000505_030 |
0.2185458528 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_078954_020 |
0.2237314252 |
- |
- |
PREDICTED: cytochrome P450 76A1-like [Jatropha curcas] |
| 7 |
Hb_012633_010 |
0.2240754871 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 49 [Jatropha curcas] |
| 8 |
Hb_002218_050 |
0.225127871 |
- |
- |
SSXT family protein isoform 2, partial [Theobroma cacao] |
| 9 |
Hb_000152_200 |
0.2256118441 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase ERL2 isoform X1 [Jatropha curcas] |
| 10 |
Hb_004542_010 |
0.2261353391 |
- |
- |
Putative chloroplast RF19 [Medicago truncatula] |
| 11 |
Hb_001433_120 |
0.2273168265 |
- |
- |
hypothetical protein CICLE_v10003574mg, partial [Citrus clementina] |
| 12 |
Hb_007157_050 |
0.2273961886 |
- |
- |
hypothetical protein RCOM_0711500 [Ricinus communis] |
| 13 |
Hb_002447_060 |
0.2279534747 |
- |
- |
PREDICTED: polyamine oxidase 1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002079_100 |
0.2296831707 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HDG5 [Jatropha curcas] |
| 15 |
Hb_003549_060 |
0.2312955452 |
- |
- |
PREDICTED: importin subunit alpha-1a-like isoform X3 [Gossypium raimondii] |
| 16 |
Hb_177533_010 |
0.231378835 |
- |
- |
Serine/threonine-protein kinase-transforming protein raf, putative [Ricinus communis] |
| 17 |
Hb_002307_040 |
0.2318507201 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 18 |
Hb_004881_040 |
0.2321681949 |
- |
- |
PREDICTED: protodermal factor 1-like [Populus euphratica] |
| 19 |
Hb_008059_040 |
0.2341006654 |
- |
- |
hypothetical protein POPTR_0019s12360g [Populus trichocarpa] |
| 20 |
Hb_001519_080 |
0.2342046326 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |