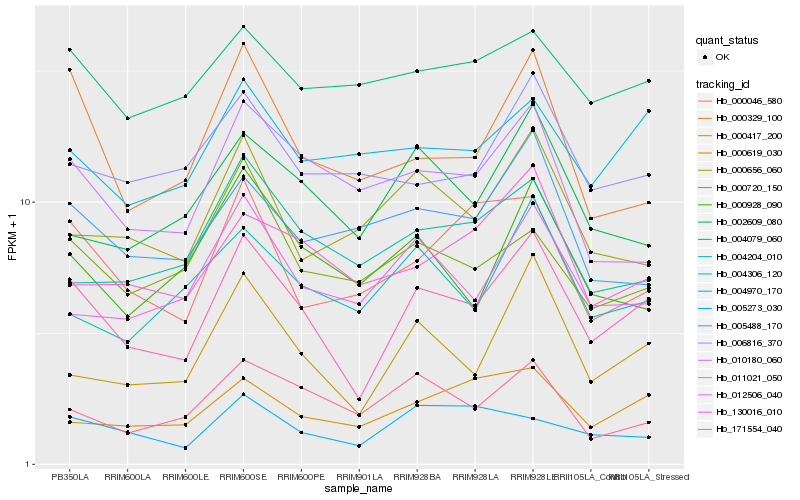
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_130016_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000619_030 |
0.1168784942 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 3 |
Hb_000329_100 |
0.1397077761 |
- |
- |
PREDICTED: uncharacterized protein LOC105643144 [Jatropha curcas] |
| 4 |
Hb_000417_200 |
0.1420828676 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: pentatricopeptide repeat-containing protein DOT4, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000046_580 |
0.1433947734 |
- |
- |
PREDICTED: uncharacterized protein LOC105631800 [Jatropha curcas] |
| 6 |
Hb_011021_050 |
0.1456532476 |
- |
- |
PREDICTED: uncharacterized protein LOC105644387 isoform X1 [Jatropha curcas] |
| 7 |
Hb_004970_170 |
0.1474841533 |
transcription factor |
TF Family: bZIP |
PREDICTED: ABSCISIC ACID-INSENSITIVE 5-like protein 5 [Jatropha curcas] |
| 8 |
Hb_002609_080 |
0.1485946686 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 7, chloroplastic-like [Jatropha curcas] |
| 9 |
Hb_012506_040 |
0.1490027822 |
transcription factor |
TF Family: DDT |
PREDICTED: uncharacterized protein LOC105637346 [Jatropha curcas] |
| 10 |
Hb_000928_090 |
0.1493435393 |
- |
- |
PREDICTED: septin and tuftelin-interacting protein 1 homolog 1 [Jatropha curcas] |
| 11 |
Hb_004306_120 |
0.1504144627 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 12 |
Hb_005273_030 |
0.15059896 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 13 |
Hb_006816_370 |
0.1509314083 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105647456 [Jatropha curcas] |
| 14 |
Hb_000720_150 |
0.1522077256 |
- |
- |
PREDICTED: uncharacterized protein LOC105645849 isoform X3 [Jatropha curcas] |
| 15 |
Hb_010180_060 |
0.1527183002 |
- |
- |
PREDICTED: uncharacterized protein LOC105648049 [Jatropha curcas] |
| 16 |
Hb_004204_010 |
0.1533272416 |
- |
- |
PREDICTED: uncharacterized protein LOC105640608 [Jatropha curcas] |
| 17 |
Hb_000656_060 |
0.1535976041 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 18 |
Hb_004079_060 |
0.1540081889 |
- |
- |
hypothetical protein JCGZ_07228 [Jatropha curcas] |
| 19 |
Hb_171554_040 |
0.1544217678 |
- |
- |
PREDICTED: phytanoyl-CoA dioxygenase [Populus euphratica] |
| 20 |
Hb_005488_170 |
0.1555237874 |
- |
- |
PREDICTED: septin and tuftelin-interacting protein 1 homolog 1 [Jatropha curcas] |