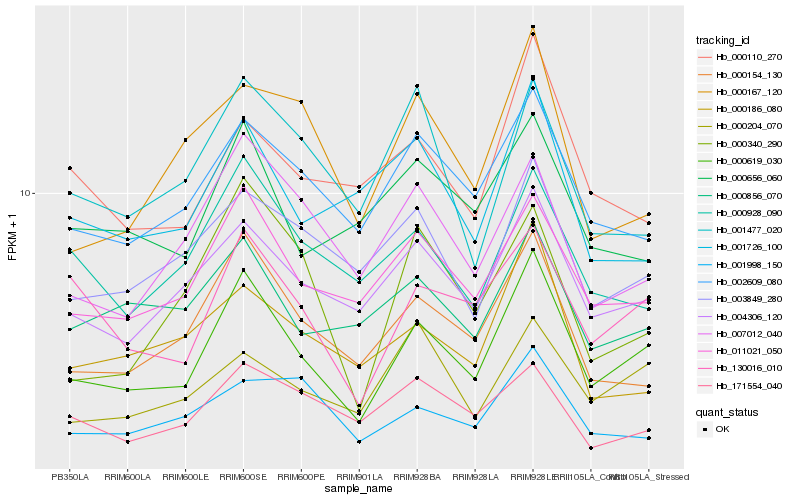
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000619_030 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_011021_050 |
0.1138497568 |
- |
- |
PREDICTED: uncharacterized protein LOC105644387 isoform X1 [Jatropha curcas] |
| 3 |
Hb_004306_120 |
0.1160996599 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 4 |
Hb_130016_010 |
0.1168784942 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000154_130 |
0.1205295347 |
- |
- |
PREDICTED: flowering time control protein FPA isoform X1 [Jatropha curcas] |
| 6 |
Hb_002609_080 |
0.1262615038 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 7, chloroplastic-like [Jatropha curcas] |
| 7 |
Hb_007012_040 |
0.1362733575 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 44 [Jatropha curcas] |
| 8 |
Hb_003849_280 |
0.1376015197 |
- |
- |
PREDICTED: uncharacterized protein LOC105644977 [Jatropha curcas] |
| 9 |
Hb_000928_090 |
0.1389103668 |
- |
- |
PREDICTED: septin and tuftelin-interacting protein 1 homolog 1 [Jatropha curcas] |
| 10 |
Hb_000856_070 |
0.1401681291 |
- |
- |
PREDICTED: formin-like protein 5 [Jatropha curcas] |
| 11 |
Hb_000204_070 |
0.1415633821 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g09900-like [Jatropha curcas] |
| 12 |
Hb_000656_060 |
0.1432822523 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 13 |
Hb_000110_270 |
0.1441782677 |
- |
- |
prp4, putative [Ricinus communis] |
| 14 |
Hb_001726_100 |
0.1453052251 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 19 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000186_080 |
0.1465825942 |
- |
- |
PREDICTED: protein unc-45 homolog B-like [Gossypium raimondii] |
| 16 |
Hb_000167_120 |
0.1474093715 |
- |
- |
PREDICTED: U-box domain-containing protein 14 [Jatropha curcas] |
| 17 |
Hb_171554_040 |
0.1489189376 |
- |
- |
PREDICTED: phytanoyl-CoA dioxygenase [Populus euphratica] |
| 18 |
Hb_001998_150 |
0.1494134305 |
- |
- |
hypothetical protein JCGZ_09140 [Jatropha curcas] |
| 19 |
Hb_000340_290 |
0.1509610464 |
- |
- |
PREDICTED: piezo-type mechanosensitive ion channel homolog [Jatropha curcas] |
| 20 |
Hb_001477_020 |
0.1514936952 |
transcription factor |
TF Family: HB |
PREDICTED: uncharacterized protein LOC105646886 isoform X3 [Jatropha curcas] |