| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
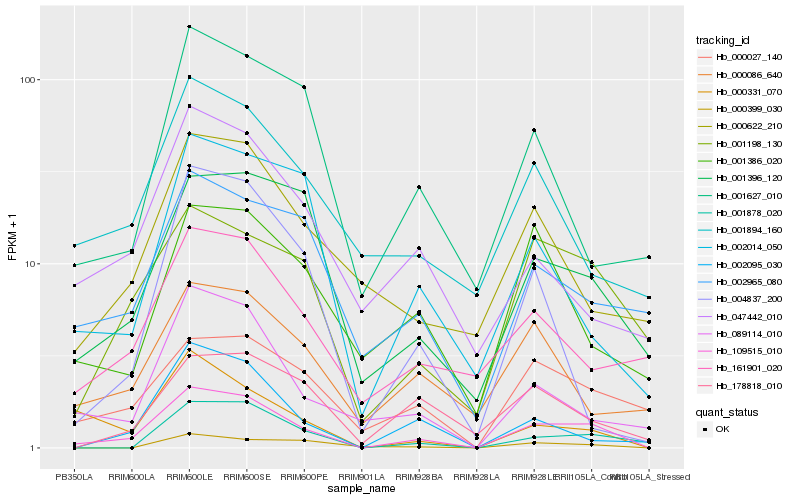
Hb_109515_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_001396_120 |
0.2214009177 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 16 [Jatropha curcas] |
| 3 |
Hb_089114_010 |
0.2234573244 |
- |
- |
PREDICTED: uncharacterized protein LOC105646926 [Jatropha curcas] |
| 4 |
Hb_001198_130 |
0.2258958222 |
- |
- |
PREDICTED: potassium channel AKT2/3 [Jatropha curcas] |
| 5 |
Hb_001878_020 |
0.2284458647 |
- |
- |
PREDICTED: uncharacterized protein LOC105628311 [Jatropha curcas] |
| 6 |
Hb_002095_030 |
0.2305752973 |
- |
- |
hypothetical protein MIMGU_mgv1a021906mg, partial [Erythranthe guttata] |
| 7 |
Hb_000331_070 |
0.2306541001 |
- |
- |
PREDICTED: uncharacterized protein LOC105636644 [Jatropha curcas] |
| 8 |
Hb_000027_140 |
0.231096771 |
- |
- |
PREDICTED: uncharacterized protein LOC105642063 [Jatropha curcas] |
| 9 |
Hb_002014_050 |
0.2333057174 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK1 isoform X2 [Populus euphratica] |
| 10 |
Hb_004837_200 |
0.2382071982 |
- |
- |
hypothetical protein POPTR_0001s26450g [Populus trichocarpa] |
| 11 |
Hb_002965_080 |
0.2445850685 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 15-like [Jatropha curcas] |
| 12 |
Hb_001894_160 |
0.2459764345 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_161901_020 |
0.2467208634 |
- |
- |
PREDICTED: serine/threonine-protein kinase AFC2 isoform X2 [Vitis vinifera] |
| 14 |
Hb_001627_010 |
0.2469831965 |
- |
- |
Alpha-1,4-glucan-protein synthase [UDP-forming], putative [Ricinus communis] |
| 15 |
Hb_047442_010 |
0.2471884326 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2 [Jatropha curcas] |
| 16 |
Hb_000622_210 |
0.2497362992 |
transcription factor |
TF Family: bZIP |
PREDICTED: common plant regulatory factor 1-like isoform X3 [Jatropha curcas] |
| 17 |
Hb_178818_010 |
0.2498016566 |
- |
- |
CRINKLY4 related 3, putative [Theobroma cacao] |
| 18 |
Hb_000399_030 |
0.2510988805 |
- |
- |
PREDICTED: uncharacterized protein LOC105634579 [Jatropha curcas] |
| 19 |
Hb_001386_020 |
0.251649078 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000086_640 |
0.2522483609 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |