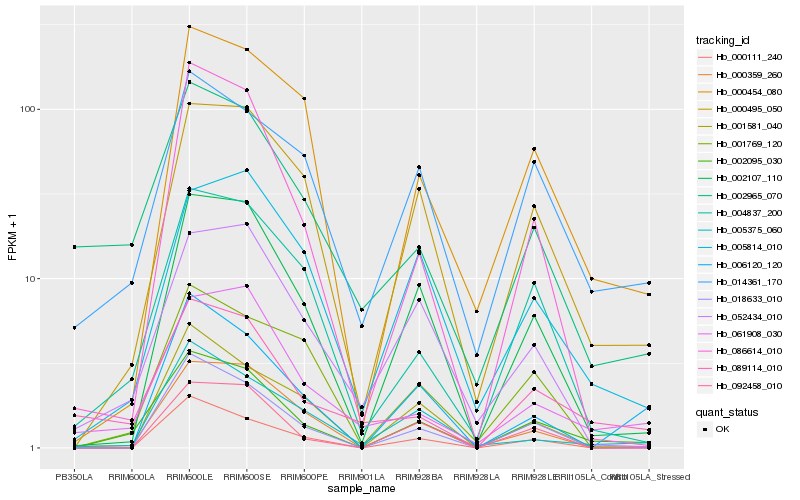
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002095_030 |
0.0 |
- |
- |
hypothetical protein MIMGU_mgv1a021906mg, partial [Erythranthe guttata] |
| 2 |
Hb_000495_050 |
0.161941293 |
- |
- |
hypothetical protein JCGZ_25174 [Jatropha curcas] |
| 3 |
Hb_002107_110 |
0.164144867 |
- |
- |
PREDICTED: probable cinnamyl alcohol dehydrogenase 6 isoform X1 [Jatropha curcas] |
| 4 |
Hb_004837_200 |
0.1713181297 |
- |
- |
hypothetical protein POPTR_0001s26450g [Populus trichocarpa] |
| 5 |
Hb_000359_260 |
0.1820309064 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 [Jatropha curcas] |
| 6 |
Hb_014361_170 |
0.1843958211 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 37 [Jatropha curcas] |
| 7 |
Hb_018633_010 |
0.1859144974 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 8 |
Hb_000454_080 |
0.1865185989 |
- |
- |
PREDICTED: arginine decarboxylase [Jatropha curcas] |
| 9 |
Hb_086614_010 |
0.1873088937 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein HD1-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_089114_010 |
0.1922744747 |
- |
- |
PREDICTED: uncharacterized protein LOC105646926 [Jatropha curcas] |
| 11 |
Hb_061908_030 |
0.1925483131 |
- |
- |
PREDICTED: putative disease resistance protein At3g14460 [Jatropha curcas] |
| 12 |
Hb_000111_240 |
0.1940816228 |
- |
- |
PREDICTED: glutamate receptor 2.8-like [Elaeis guineensis] |
| 13 |
Hb_002965_070 |
0.194586821 |
- |
- |
PREDICTED: extensin [Jatropha curcas] |
| 14 |
Hb_005375_060 |
0.1998914758 |
- |
- |
PREDICTED: probably inactive receptor-like protein kinase At2g46850 [Jatropha curcas] |
| 15 |
Hb_001581_040 |
0.2001919962 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 16 |
Hb_005814_010 |
0.2003663884 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 17 |
Hb_092458_010 |
0.2017145144 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 18 |
Hb_052434_010 |
0.2043819182 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like serine/threonine-protein kinase At3g14840 [Sesamum indicum] |
| 19 |
Hb_001769_120 |
0.2047943953 |
- |
- |
PREDICTED: tetratricopeptide repeat protein 7A [Jatropha curcas] |
| 20 |
Hb_006120_120 |
0.2080724604 |
- |
- |
conserved hypothetical protein [Ricinus communis] |