| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
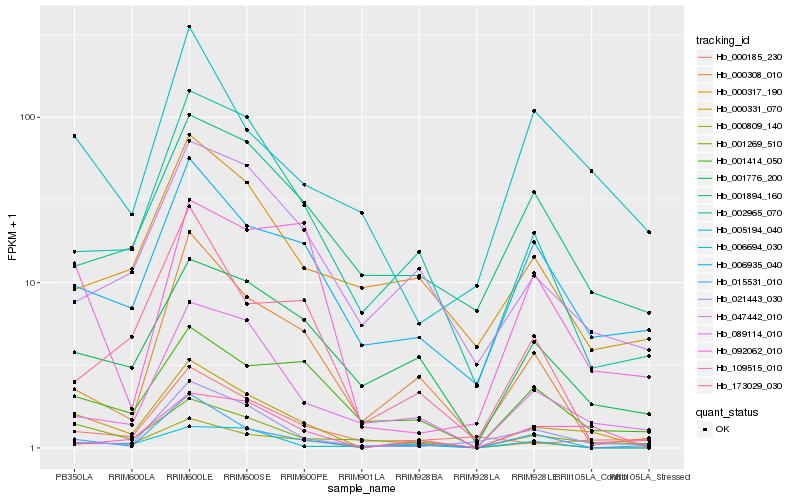
Hb_000331_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636644 [Jatropha curcas] |
| 2 |
Hb_001269_510 |
0.2065061488 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 3 |
Hb_002965_070 |
0.2196605686 |
- |
- |
PREDICTED: extensin [Jatropha curcas] |
| 4 |
Hb_089114_010 |
0.2270163764 |
- |
- |
PREDICTED: uncharacterized protein LOC105646926 [Jatropha curcas] |
| 5 |
Hb_109515_010 |
0.2306541001 |
- |
- |
- |
| 6 |
Hb_006935_040 |
0.230904867 |
- |
- |
PREDICTED: uncharacterized protein LOC105629236 [Jatropha curcas] |
| 7 |
Hb_001776_200 |
0.2379373669 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 2 isoform X2 [Pyrus x bretschneideri] |
| 8 |
Hb_000809_140 |
0.2391911552 |
- |
- |
hypothetical protein JCGZ_12354 [Jatropha curcas] |
| 9 |
Hb_000308_010 |
0.2447859845 |
- |
- |
PREDICTED: uncharacterized protein LOC105639684 [Jatropha curcas] |
| 10 |
Hb_047442_010 |
0.2461712011 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2 [Jatropha curcas] |
| 11 |
Hb_001414_050 |
0.2465448225 |
- |
- |
PREDICTED: probable S-adenosylmethionine-dependent methyltransferase At5g37970 [Jatropha curcas] |
| 12 |
Hb_015531_010 |
0.2474060429 |
- |
- |
PREDICTED: uncharacterized protein LOC105636764 [Jatropha curcas] |
| 13 |
Hb_005194_040 |
0.2485835742 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 14 |
Hb_173029_030 |
0.2524752951 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_021443_030 |
0.2533096537 |
- |
- |
PREDICTED: uncharacterized protein LOC105648566 [Jatropha curcas] |
| 16 |
Hb_000185_230 |
0.2534163301 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 17 |
Hb_001894_160 |
0.2573760301 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000317_190 |
0.2585063195 |
- |
- |
PREDICTED: diacylglycerol kinase 5-like [Jatropha curcas] |
| 19 |
Hb_092062_010 |
0.2621001484 |
- |
- |
hypothetical protein JCGZ_25129 [Jatropha curcas] |
| 20 |
Hb_006694_030 |
0.2629006808 |
- |
- |
- |