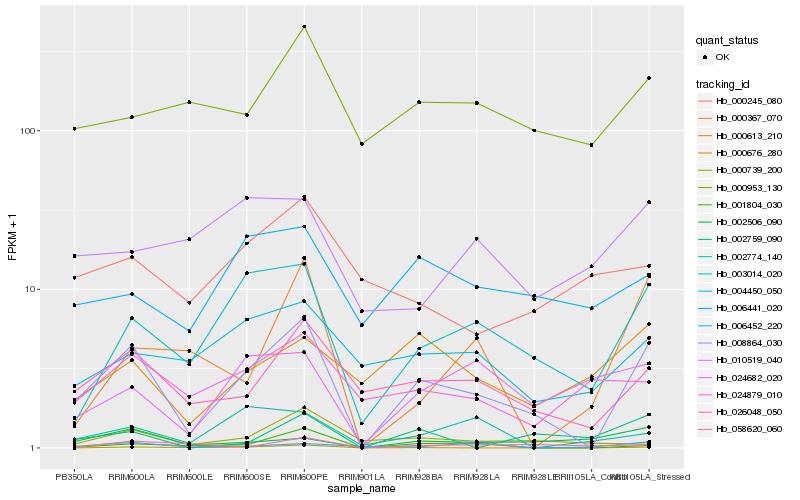
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008864_030 |
0.0 |
- |
- |
- |
| 2 |
Hb_003014_020 |
0.2490442153 |
- |
- |
electron transporter, putative [Ricinus communis] |
| 3 |
Hb_026048_050 |
0.2510052032 |
- |
- |
PREDICTED: probable membrane-associated kinase regulator 2 [Jatropha curcas] |
| 4 |
Hb_001804_030 |
0.3093379037 |
- |
- |
kinase, putative [Ricinus communis] |
| 5 |
Hb_004450_050 |
0.312330608 |
- |
- |
PREDICTED: histone chaperone ASF1B [Populus euphratica] |
| 6 |
Hb_024682_020 |
0.3148999531 |
- |
- |
hypothetical protein POPTR_0001s12710g [Populus trichocarpa] |
| 7 |
Hb_002506_090 |
0.3156243264 |
- |
- |
- |
| 8 |
Hb_000953_130 |
0.3160072816 |
- |
- |
PREDICTED: fructose-bisphosphate aldolase cytoplasmic isozyme [Jatropha curcas] |
| 9 |
Hb_000613_210 |
0.3181864037 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 10 |
Hb_006441_020 |
0.3184917773 |
- |
- |
PREDICTED: cadmium/zinc-transporting ATPase HMA3-like isoform X2 [Populus euphratica] |
| 11 |
Hb_024879_010 |
0.3215870201 |
- |
- |
unknown [Glycine max] |
| 12 |
Hb_000676_280 |
0.32576861 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 13 |
Hb_000739_200 |
0.3265912868 |
transcription factor |
TF Family: LOB |
hypothetical protein CICLE_v10021992mg [Citrus clementina] |
| 14 |
Hb_000367_070 |
0.330252297 |
- |
- |
hypothetical protein JCGZ_11697 [Jatropha curcas] |
| 15 |
Hb_002759_090 |
0.3307234138 |
- |
- |
- |
| 16 |
Hb_006452_220 |
0.3325519057 |
- |
- |
PREDICTED: uncharacterized protein LOC105635043 [Jatropha curcas] |
| 17 |
Hb_002774_140 |
0.3353902987 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_058620_060 |
0.3360579607 |
- |
- |
uncharacterized protein LOC105646159 precursor [Jatropha curcas] |
| 19 |
Hb_000245_080 |
0.3410516872 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-15 [Jatropha curcas] |
| 20 |
Hb_010519_040 |
0.344603626 |
- |
- |
PREDICTED: uncharacterized protein LOC105643567 [Jatropha curcas] |