| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
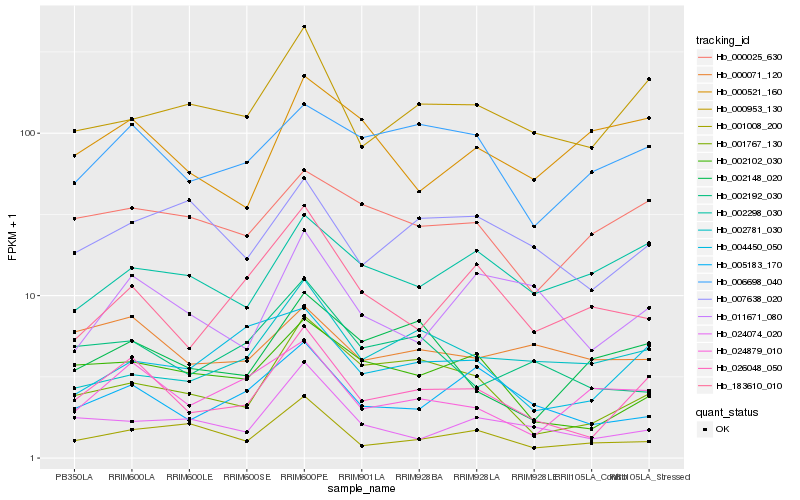
Hb_026048_050 |
0.0 |
- |
- |
PREDICTED: probable membrane-associated kinase regulator 2 [Jatropha curcas] |
| 2 |
Hb_000953_130 |
0.1649704736 |
- |
- |
PREDICTED: fructose-bisphosphate aldolase cytoplasmic isozyme [Jatropha curcas] |
| 3 |
Hb_001767_130 |
0.1706267272 |
- |
- |
PREDICTED: beta-hexosaminidase 1 [Jatropha curcas] |
| 4 |
Hb_002102_030 |
0.172978027 |
- |
- |
- |
| 5 |
Hb_006698_040 |
0.1786793077 |
- |
- |
bHLH1 transcription factor [Hevea brasiliensis] |
| 6 |
Hb_005183_170 |
0.1798282683 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g04860 [Jatropha curcas] |
| 7 |
Hb_011671_080 |
0.1877728177 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like protein kinase TDR [Jatropha curcas] |
| 8 |
Hb_024879_010 |
0.1892025243 |
- |
- |
unknown [Glycine max] |
| 9 |
Hb_004450_050 |
0.1908504084 |
- |
- |
PREDICTED: histone chaperone ASF1B [Populus euphratica] |
| 10 |
Hb_000025_630 |
0.1921785231 |
- |
- |
PREDICTED: uncharacterized protein LOC105635952 [Jatropha curcas] |
| 11 |
Hb_001008_200 |
0.192856702 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC103961552 [Pyrus x bretschneideri] |
| 12 |
Hb_024074_020 |
0.1944545481 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g09600 [Jatropha curcas] |
| 13 |
Hb_000071_120 |
0.1999587239 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 4 [Jatropha curcas] |
| 14 |
Hb_002148_020 |
0.2009642287 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 15 |
Hb_002192_030 |
0.2018399072 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_007638_020 |
0.2018478927 |
- |
- |
PREDICTED: low-temperature-induced cysteine proteinase-like [Jatropha curcas] |
| 17 |
Hb_002298_030 |
0.203101913 |
- |
- |
PREDICTED: coenzyme Q-binding protein COQ10 homolog, mitochondrial [Jatropha curcas] |
| 18 |
Hb_183610_010 |
0.2067606307 |
- |
- |
PREDICTED: uncharacterized protein LOC105635777 [Jatropha curcas] |
| 19 |
Hb_002781_030 |
0.2072200592 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 20 |
Hb_000521_160 |
0.2077371505 |
- |
- |
conserved hypothetical protein [Ricinus communis] |