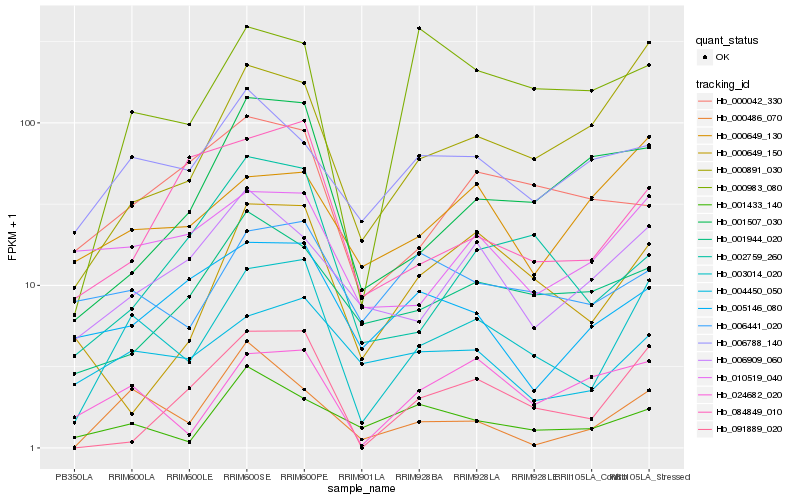
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003014_020 |
0.0 |
- |
- |
electron transporter, putative [Ricinus communis] |
| 2 |
Hb_004450_050 |
0.2113426637 |
- |
- |
PREDICTED: histone chaperone ASF1B [Populus euphratica] |
| 3 |
Hb_024682_020 |
0.2126636815 |
- |
- |
hypothetical protein POPTR_0001s12710g [Populus trichocarpa] |
| 4 |
Hb_000891_030 |
0.2153943089 |
- |
- |
PREDICTED: uncharacterized protein At4g22758 [Jatropha curcas] |
| 5 |
Hb_010519_040 |
0.223887925 |
- |
- |
PREDICTED: uncharacterized protein LOC105643567 [Jatropha curcas] |
| 6 |
Hb_091889_020 |
0.22510219 |
- |
- |
- |
| 7 |
Hb_006441_020 |
0.2310831445 |
- |
- |
PREDICTED: cadmium/zinc-transporting ATPase HMA3-like isoform X2 [Populus euphratica] |
| 8 |
Hb_006909_060 |
0.2328191822 |
- |
- |
PREDICTED: eukaryotic peptide chain release factor subunit 1-3-like [Solanum lycopersicum] |
| 9 |
Hb_006788_140 |
0.2376076819 |
- |
- |
PREDICTED: uncharacterized protein LOC105649915 [Jatropha curcas] |
| 10 |
Hb_001507_030 |
0.2382224833 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 2 [Jatropha curcas] |
| 11 |
Hb_000486_070 |
0.2383687644 |
- |
- |
hypothetical protein JCGZ_25069 [Jatropha curcas] |
| 12 |
Hb_000983_080 |
0.2405849366 |
transcription factor |
TF Family: bZIP |
PREDICTED: ocs element-binding factor 1-like [Jatropha curcas] |
| 13 |
Hb_000649_130 |
0.2410977765 |
- |
- |
Polcalcin Jun o, putative [Ricinus communis] |
| 14 |
Hb_001944_020 |
0.2416329626 |
- |
- |
PREDICTED: uncharacterized protein LOC105636535 [Jatropha curcas] |
| 15 |
Hb_000649_150 |
0.2419554573 |
- |
- |
PREDICTED: uncharacterized protein LOC105634115 [Jatropha curcas] |
| 16 |
Hb_084849_010 |
0.242014473 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002759_260 |
0.2424397571 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 9 [Jatropha curcas] |
| 18 |
Hb_001433_140 |
0.2440075709 |
- |
- |
PREDICTED: phospholipase D p1 [Jatropha curcas] |
| 19 |
Hb_005146_080 |
0.244113683 |
- |
- |
PREDICTED: uncharacterized protein LOC105636704 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000042_330 |
0.2453467601 |
- |
- |
conserved hypothetical protein [Ricinus communis] |