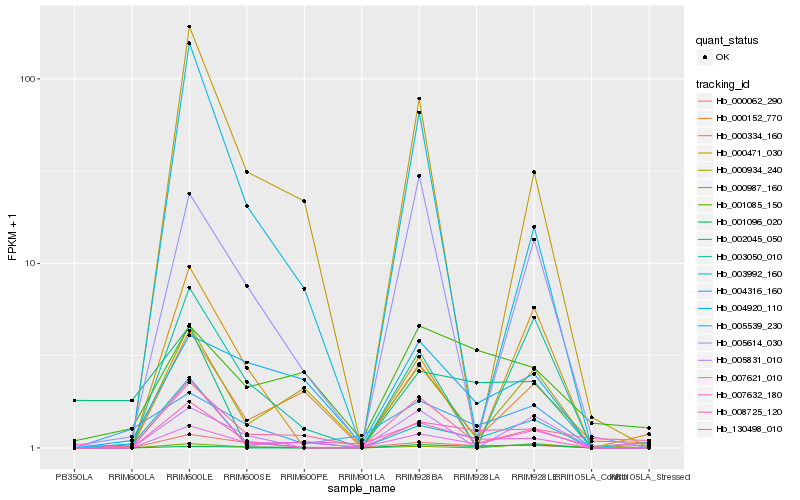
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008725_120 |
0.0 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 16-like [Populus euphratica] |
| 2 |
Hb_000062_290 |
0.2598380904 |
desease resistance |
Gene Name: ABC_membrane |
hypothetical protein POPTR_0008s17960g [Populus trichocarpa] |
| 3 |
Hb_003992_160 |
0.2725310967 |
- |
- |
PREDICTED: cucumber peeling cupredoxin-like [Jatropha curcas] |
| 4 |
Hb_007632_180 |
0.3012640551 |
- |
- |
Pectate lyase precursor, putative [Ricinus communis] |
| 5 |
Hb_005539_230 |
0.3039349403 |
- |
- |
hypothetical protein L484_027799 [Morus notabilis] |
| 6 |
Hb_005831_010 |
0.305642073 |
- |
- |
hypothetical protein JCGZ_04003 [Jatropha curcas] |
| 7 |
Hb_003050_010 |
0.3169058187 |
- |
- |
PREDICTED: VQ motif-containing protein 1-like [Jatropha curcas] |
| 8 |
Hb_000334_160 |
0.3232029111 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_004316_160 |
0.3249970096 |
- |
- |
hypothetical protein CISIN_1g047906mg, partial [Citrus sinensis] |
| 10 |
Hb_005614_030 |
0.3294278723 |
- |
- |
PREDICTED: transcription factor SCREAM2-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_001096_020 |
0.3297323933 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X1 [Populus euphratica] |
| 12 |
Hb_007621_010 |
0.3324055986 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000934_240 |
0.3329446231 |
- |
- |
PREDICTED: peroxidase 13 [Jatropha curcas] |
| 14 |
Hb_000471_030 |
0.3366067416 |
- |
- |
- |
| 15 |
Hb_000152_770 |
0.3377629644 |
- |
- |
PREDICTED: dynein light chain, cytoplasmic-like [Jatropha curcas] |
| 16 |
Hb_002045_050 |
0.3388408233 |
- |
- |
- |
| 17 |
Hb_130498_010 |
0.3391471773 |
- |
- |
Laccase 17 [Theobroma cacao] |
| 18 |
Hb_004920_110 |
0.3393045294 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001085_150 |
0.3408414926 |
- |
- |
Purple acid phosphatase 15 [Theobroma cacao] |
| 20 |
Hb_000987_160 |
0.3414101403 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |