| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
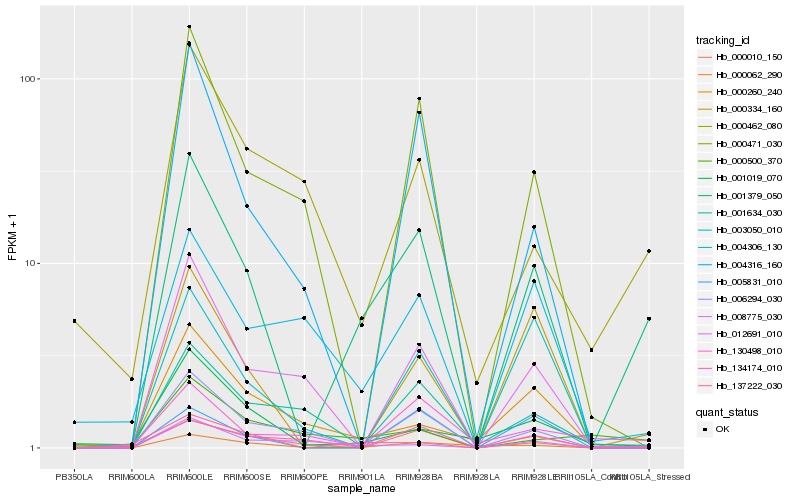
Hb_001379_050 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000062_290 |
0.2507373532 |
desease resistance |
Gene Name: ABC_membrane |
hypothetical protein POPTR_0008s17960g [Populus trichocarpa] |
| 3 |
Hb_000334_160 |
0.2571619728 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_137222_030 |
0.2822298917 |
- |
- |
- |
| 5 |
Hb_004316_160 |
0.2830196493 |
- |
- |
hypothetical protein CISIN_1g047906mg, partial [Citrus sinensis] |
| 6 |
Hb_134174_010 |
0.2884247444 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 7 |
Hb_000471_030 |
0.2939713405 |
- |
- |
- |
| 8 |
Hb_001019_070 |
0.2957278382 |
- |
- |
hypothetical protein POPTR_0007s05770g [Populus trichocarpa] |
| 9 |
Hb_003050_010 |
0.3016491069 |
- |
- |
PREDICTED: VQ motif-containing protein 1-like [Jatropha curcas] |
| 10 |
Hb_000462_080 |
0.3029523051 |
- |
- |
PREDICTED: quercetin 3-O-methyltransferase 1 [Jatropha curcas] |
| 11 |
Hb_006294_030 |
0.3032981072 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 12 |
Hb_012691_010 |
0.3064316832 |
- |
- |
hypothetical protein POPTR_0005s18750g [Populus trichocarpa] |
| 13 |
Hb_008775_030 |
0.3070490783 |
- |
- |
PREDICTED: ferric reduction oxidase 7, chloroplastic-like [Jatropha curcas] |
| 14 |
Hb_130498_010 |
0.3078538591 |
- |
- |
Laccase 17 [Theobroma cacao] |
| 15 |
Hb_001634_030 |
0.3094183513 |
- |
- |
Receptor like protein 1, putative [Theobroma cacao] |
| 16 |
Hb_005831_010 |
0.3102180803 |
- |
- |
hypothetical protein JCGZ_04003 [Jatropha curcas] |
| 17 |
Hb_004306_130 |
0.3137970889 |
- |
- |
PREDICTED: uncharacterized protein LOC105644385 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000260_240 |
0.3178327945 |
- |
- |
hypothetical protein JCGZ_00622 [Jatropha curcas] |
| 19 |
Hb_000010_150 |
0.3178519211 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RGLG2-like [Jatropha curcas] |
| 20 |
Hb_000500_370 |
0.3190507431 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |