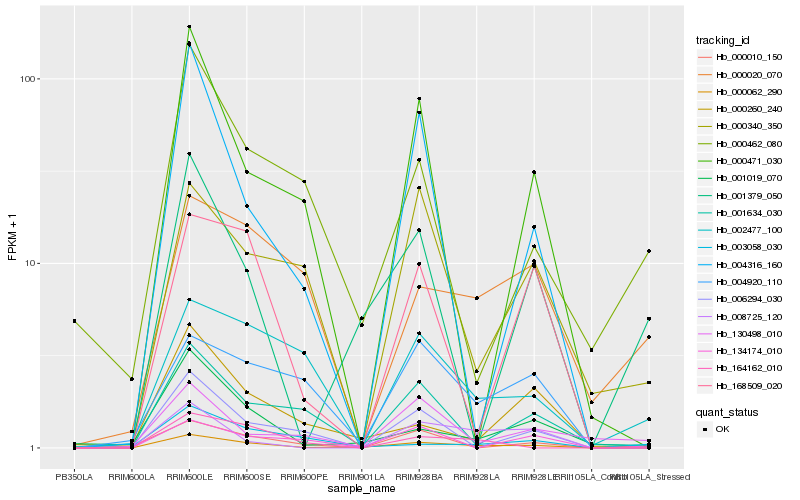
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000062_290 |
0.0 |
desease resistance |
Gene Name: ABC_membrane |
hypothetical protein POPTR_0008s17960g [Populus trichocarpa] |
| 2 |
Hb_001019_070 |
0.2366868156 |
- |
- |
hypothetical protein POPTR_0007s05770g [Populus trichocarpa] |
| 3 |
Hb_001379_050 |
0.2507373532 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001634_030 |
0.2560702757 |
- |
- |
Receptor like protein 1, putative [Theobroma cacao] |
| 5 |
Hb_002477_100 |
0.2584996588 |
- |
- |
PREDICTED: protein Brevis radix-like 2 [Jatropha curcas] |
| 6 |
Hb_008725_120 |
0.2598380904 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 16-like [Populus euphratica] |
| 7 |
Hb_000020_070 |
0.2718164325 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase NAK [Jatropha curcas] |
| 8 |
Hb_006294_030 |
0.2738916918 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 9 |
Hb_004920_110 |
0.2741129284 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_168509_020 |
0.2744274055 |
- |
- |
macrophage migration inhibitory factor family protein [Populus trichocarpa] |
| 11 |
Hb_130498_010 |
0.2752586014 |
- |
- |
Laccase 17 [Theobroma cacao] |
| 12 |
Hb_000462_080 |
0.2759854556 |
- |
- |
PREDICTED: quercetin 3-O-methyltransferase 1 [Jatropha curcas] |
| 13 |
Hb_000260_240 |
0.2760892 |
- |
- |
hypothetical protein JCGZ_00622 [Jatropha curcas] |
| 14 |
Hb_000010_150 |
0.2770750894 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RGLG2-like [Jatropha curcas] |
| 15 |
Hb_000340_350 |
0.2793061387 |
- |
- |
PREDICTED: sugar transporter ERD6-like 7 isoform X1 [Jatropha curcas] |
| 16 |
Hb_134174_010 |
0.2807671305 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 17 |
Hb_004316_160 |
0.2816529439 |
- |
- |
hypothetical protein CISIN_1g047906mg, partial [Citrus sinensis] |
| 18 |
Hb_000471_030 |
0.2823114375 |
- |
- |
- |
| 19 |
Hb_003058_030 |
0.2828940992 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 20 |
Hb_164162_010 |
0.2853785118 |
- |
- |
PREDICTED: uncharacterized protein At1g08160-like [Jatropha curcas] |