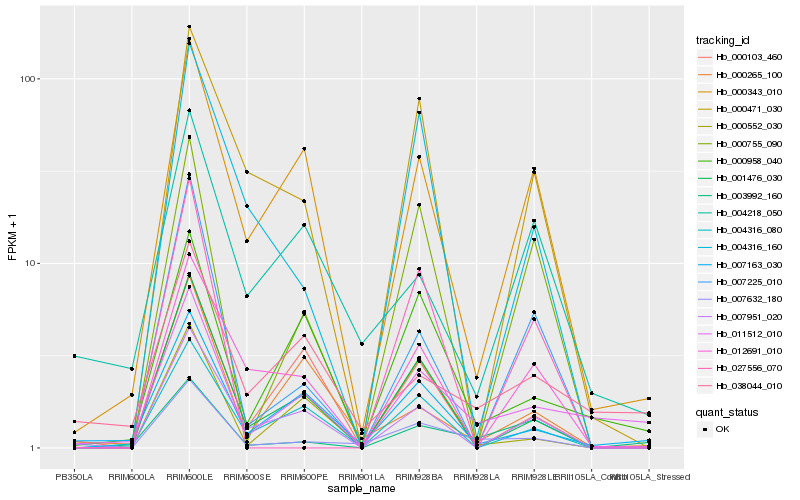
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007632_180 |
0.0 |
- |
- |
Pectate lyase precursor, putative [Ricinus communis] |
| 2 |
Hb_001476_030 |
0.2449619888 |
- |
- |
Cationic peroxidase 2 precursor, putative [Ricinus communis] |
| 3 |
Hb_007163_030 |
0.2453128443 |
- |
- |
hypothetical protein RCOM_0187400 [Ricinus communis] |
| 4 |
Hb_011512_010 |
0.2579043463 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 5 |
Hb_003992_160 |
0.2635652166 |
- |
- |
PREDICTED: cucumber peeling cupredoxin-like [Jatropha curcas] |
| 6 |
Hb_004316_160 |
0.2642534786 |
- |
- |
hypothetical protein CISIN_1g047906mg, partial [Citrus sinensis] |
| 7 |
Hb_007225_010 |
0.2707984259 |
- |
- |
PREDICTED: glycerol-3-phosphate 2-O-acyltransferase 6 [Populus euphratica] |
| 8 |
Hb_004316_080 |
0.2711895235 |
- |
- |
PREDICTED: uncharacterized protein LOC104602212 [Nelumbo nucifera] |
| 9 |
Hb_007951_020 |
0.2726792842 |
- |
- |
cationic amino acid transporter 5 family protein [Populus trichocarpa] |
| 10 |
Hb_000755_090 |
0.2752474248 |
- |
- |
transferase, putative [Ricinus communis] |
| 11 |
Hb_000343_010 |
0.2780920692 |
- |
- |
F3F9.11 [Arabidopsis thaliana] |
| 12 |
Hb_000471_030 |
0.2794266321 |
- |
- |
- |
| 13 |
Hb_000958_040 |
0.2855129565 |
- |
- |
PREDICTED: DNA topoisomerase 2 [Jatropha curcas] |
| 14 |
Hb_000552_030 |
0.2861411688 |
- |
- |
glucose-6-phosphate 1-dehydrogenase, putative [Ricinus communis] |
| 15 |
Hb_004218_050 |
0.2863615252 |
- |
- |
annexin [Manihot esculenta] |
| 16 |
Hb_027556_070 |
0.2876220906 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_038044_010 |
0.2876747409 |
- |
- |
12-oxophytodienoate reductase 2 [Theobroma cacao] |
| 18 |
Hb_012691_010 |
0.288901555 |
- |
- |
hypothetical protein POPTR_0005s18750g [Populus trichocarpa] |
| 19 |
Hb_000265_100 |
0.2895312256 |
- |
- |
PREDICTED: uncharacterized protein LOC105643473 [Jatropha curcas] |
| 20 |
Hb_000103_460 |
0.2912341114 |
- |
- |
PREDICTED: calmodulin-like protein 11 [Jatropha curcas] |