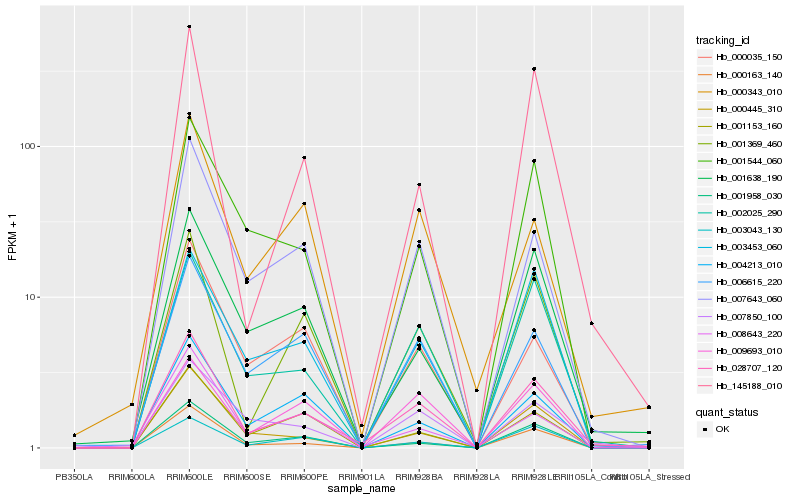
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_028707_120 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105645799 isoform X2 [Jatropha curcas] |
| 2 |
Hb_001369_460 |
0.1308459709 |
- |
- |
vacuolar invertase [Manihot esculenta] |
| 3 |
Hb_000163_140 |
0.1308905525 |
- |
- |
PREDICTED: caffeoylshikimate esterase [Jatropha curcas] |
| 4 |
Hb_002025_290 |
0.1340161824 |
transcription factor |
TF Family: G2-like |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_007643_060 |
0.1348711462 |
- |
- |
- |
| 6 |
Hb_006615_220 |
0.1478704117 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor BIM1 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000445_310 |
0.1506927267 |
- |
- |
hypothetical protein B456_011G218000 [Gossypium raimondii] |
| 8 |
Hb_001638_190 |
0.1515735233 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_001958_030 |
0.1532532052 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 10 |
Hb_145188_010 |
0.1546315957 |
- |
- |
PREDICTED: endochitinase PR4-like [Jatropha curcas] |
| 11 |
Hb_003453_060 |
0.1577061349 |
- |
- |
PREDICTED: nudix hydrolase 13, mitochondrial [Jatropha curcas] |
| 12 |
Hb_003043_130 |
0.1630238527 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor CRF2-like [Jatropha curcas] |
| 13 |
Hb_001544_060 |
0.1634367623 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB4 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000343_010 |
0.1637925457 |
- |
- |
F3F9.11 [Arabidopsis thaliana] |
| 15 |
Hb_001153_160 |
0.1639396819 |
- |
- |
molybdopterin cofactor sulfurase, putative [Ricinus communis] |
| 16 |
Hb_007850_100 |
0.1649200448 |
- |
- |
PREDICTED: uncharacterized protein LOC105640661 [Jatropha curcas] |
| 17 |
Hb_008643_220 |
0.165638803 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53440 [Jatropha curcas] |
| 18 |
Hb_004213_010 |
0.1669043742 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 19 |
Hb_000035_150 |
0.1673387918 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 40 [Jatropha curcas] |
| 20 |
Hb_009693_010 |
0.1681282141 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK15 [Gossypium raimondii] |