| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
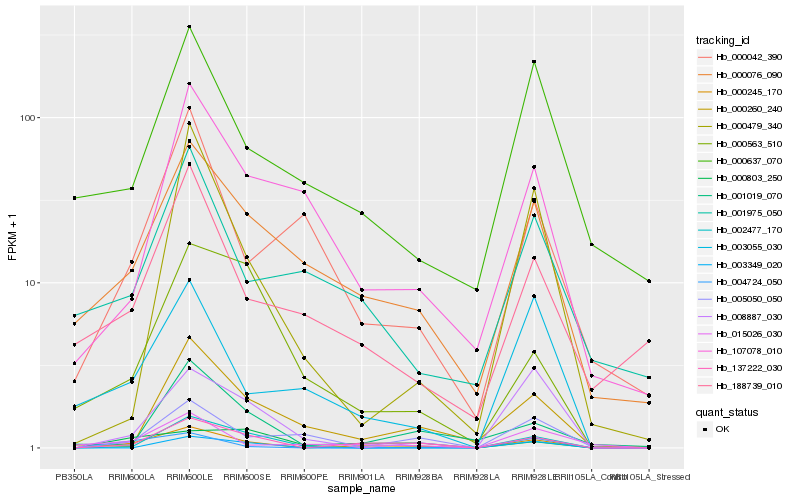
Hb_000245_170 |
0.0 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 2 |
Hb_137222_030 |
0.1301908608 |
- |
- |
- |
| 3 |
Hb_000076_090 |
0.2539225015 |
- |
- |
protein with unknown function [Ricinus communis] |
| 4 |
Hb_001019_070 |
0.2853311906 |
- |
- |
hypothetical protein POPTR_0007s05770g [Populus trichocarpa] |
| 5 |
Hb_004724_050 |
0.2888126425 |
transcription factor |
TF Family: bZIP |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000479_340 |
0.2902767459 |
- |
- |
PREDICTED: uncharacterized protein LOC105644112 [Jatropha curcas] |
| 7 |
Hb_008887_030 |
0.2908975503 |
- |
- |
PREDICTED: uncharacterized protein LOC105650448 [Jatropha curcas] |
| 8 |
Hb_015026_030 |
0.2914621338 |
- |
- |
hypothetical protein POPTR_0011s04350g [Populus trichocarpa] |
| 9 |
Hb_000260_240 |
0.2923195491 |
- |
- |
hypothetical protein JCGZ_00622 [Jatropha curcas] |
| 10 |
Hb_003055_030 |
0.2955864299 |
- |
- |
PREDICTED: glycine-rich cell wall structural protein 2 [Jatropha curcas] |
| 11 |
Hb_188739_010 |
0.2960213548 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 12 |
Hb_002477_170 |
0.2965991057 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_005050_050 |
0.2971890183 |
- |
- |
PREDICTED: wall-associated receptor kinase 5-like [Glycine max] |
| 14 |
Hb_000637_070 |
0.2981835105 |
- |
- |
PREDICTED: uncharacterized protein LOC105650668 [Jatropha curcas] |
| 15 |
Hb_107078_010 |
0.2982265785 |
- |
- |
- |
| 16 |
Hb_003349_020 |
0.3026411001 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein At4g27190-like [Beta vulgaris subsp. vulgaris] |
| 17 |
Hb_000803_250 |
0.3046416978 |
- |
- |
PREDICTED: uncharacterized protein LOC105648317 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000042_390 |
0.3053736868 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 7 [Jatropha curcas] |
| 19 |
Hb_000563_510 |
0.3070058165 |
- |
- |
epoxide hydrolase, putative [Ricinus communis] |
| 20 |
Hb_001975_050 |
0.3100720728 |
- |
- |
Tyrosine-specific transport protein, putative [Ricinus communis] |