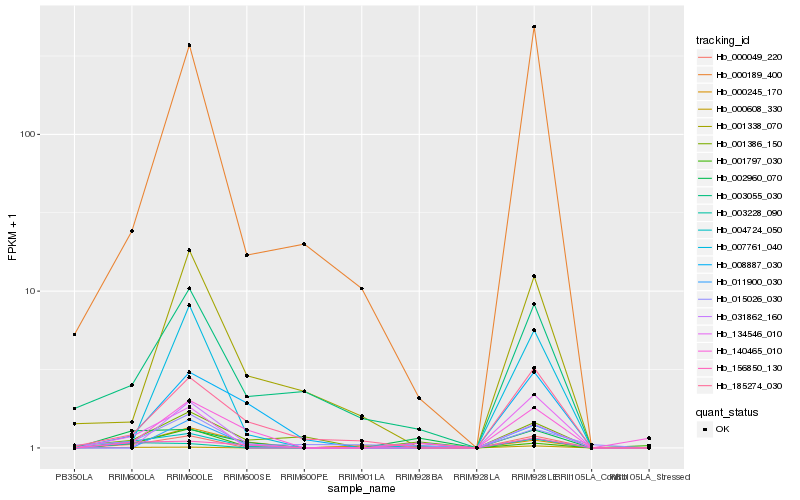
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004724_050 |
0.0 |
transcription factor |
TF Family: bZIP |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001797_030 |
0.2586182077 |
- |
- |
PREDICTED: uncharacterized protein LOC104234330 [Nicotiana sylvestris] |
| 3 |
Hb_000049_220 |
0.2710608195 |
- |
- |
ammonium transporter, putative [Ricinus communis] |
| 4 |
Hb_003228_090 |
0.2740036614 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105637258 [Jatropha curcas] |
| 5 |
Hb_000245_170 |
0.2888126425 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 6 |
Hb_134546_010 |
0.2922741285 |
- |
- |
PREDICTED: uncharacterized protein LOC105107372 [Populus euphratica] |
| 7 |
Hb_000608_330 |
0.3016736756 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 8 |
Hb_002960_070 |
0.303224749 |
- |
- |
- |
| 9 |
Hb_003055_030 |
0.3063258071 |
- |
- |
PREDICTED: glycine-rich cell wall structural protein 2 [Jatropha curcas] |
| 10 |
Hb_185274_030 |
0.3073469935 |
- |
- |
hypothetical protein JCGZ_15170 [Jatropha curcas] |
| 11 |
Hb_007761_040 |
0.3166019205 |
- |
- |
carbonyl reductase, putative [Ricinus communis] |
| 12 |
Hb_140465_010 |
0.3176804466 |
- |
- |
PREDICTED: MLP-like protein 423 [Jatropha curcas] |
| 13 |
Hb_000189_400 |
0.3255490675 |
- |
- |
hypothetical protein CathTA2_0066 [Caldalkalibacillus thermarum TA2.A1] |
| 14 |
Hb_008887_030 |
0.3267410098 |
- |
- |
PREDICTED: uncharacterized protein LOC105650448 [Jatropha curcas] |
| 15 |
Hb_015026_030 |
0.3290068301 |
- |
- |
hypothetical protein POPTR_0011s04350g [Populus trichocarpa] |
| 16 |
Hb_001338_070 |
0.3320547285 |
- |
- |
ORF126 [Jatropha curcas] |
| 17 |
Hb_011900_030 |
0.335303082 |
- |
- |
hypothetical protein JCGZ_13686 [Jatropha curcas] |
| 18 |
Hb_031862_160 |
0.3391397953 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 19 |
Hb_156850_130 |
0.3392764883 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001386_150 |
0.3437808345 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |