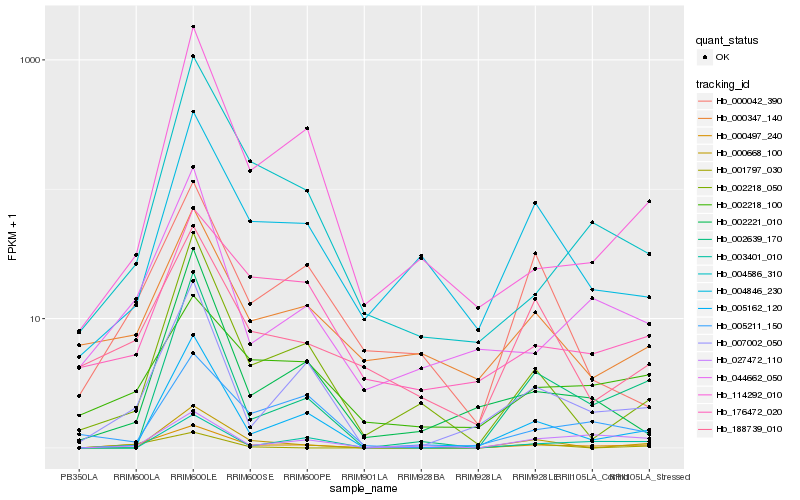
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000497_240 |
0.0 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein indeterminate-domain 2-like [Jatropha curcas] |
| 2 |
Hb_007002_050 |
0.1983160794 |
- |
- |
PREDICTED: uncharacterized protein LOC105642607 [Jatropha curcas] |
| 3 |
Hb_005162_120 |
0.2220487576 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |
| 4 |
Hb_044662_050 |
0.2310467608 |
- |
- |
hypothetical protein POPTR_0002s08440g [Populus trichocarpa] |
| 5 |
Hb_002639_170 |
0.2391235705 |
- |
- |
carboxy-lyase, putative [Ricinus communis] |
| 6 |
Hb_002218_100 |
0.2455250089 |
- |
- |
PREDICTED: uncharacterized protein At2g34160 [Jatropha curcas] |
| 7 |
Hb_004586_310 |
0.2467755712 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002218_050 |
0.2505403749 |
- |
- |
SSXT family protein isoform 2, partial [Theobroma cacao] |
| 9 |
Hb_188739_010 |
0.254589245 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 10 |
Hb_027472_110 |
0.2577606446 |
- |
- |
Patellin-4, putative [Ricinus communis] |
| 11 |
Hb_003401_010 |
0.2618085641 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 12 |
Hb_004846_230 |
0.2646025343 |
- |
- |
Chaperone protein dnaJ 8, chloroplast precursor, putative [Ricinus communis] |
| 13 |
Hb_001797_030 |
0.2648576742 |
- |
- |
PREDICTED: uncharacterized protein LOC104234330 [Nicotiana sylvestris] |
| 14 |
Hb_176472_020 |
0.2664935803 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000347_140 |
0.2667169585 |
- |
- |
PREDICTED: protein FLUORESCENT IN BLUE LIGHT, chloroplastic isoform X2 [Jatropha curcas] |
| 16 |
Hb_114292_010 |
0.2675263649 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002221_010 |
0.2706277379 |
- |
- |
PREDICTED: adenylyl-sulfate kinase 3-like [Jatropha curcas] |
| 18 |
Hb_000042_390 |
0.2720731624 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 7 [Jatropha curcas] |
| 19 |
Hb_000668_100 |
0.2720841776 |
- |
- |
PREDICTED: non-specific lipid-transfer protein-like protein At2g13820 [Jatropha curcas] |
| 20 |
Hb_005211_150 |
0.2729636945 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HBP-1b(c38) isoform X1 [Jatropha curcas] |