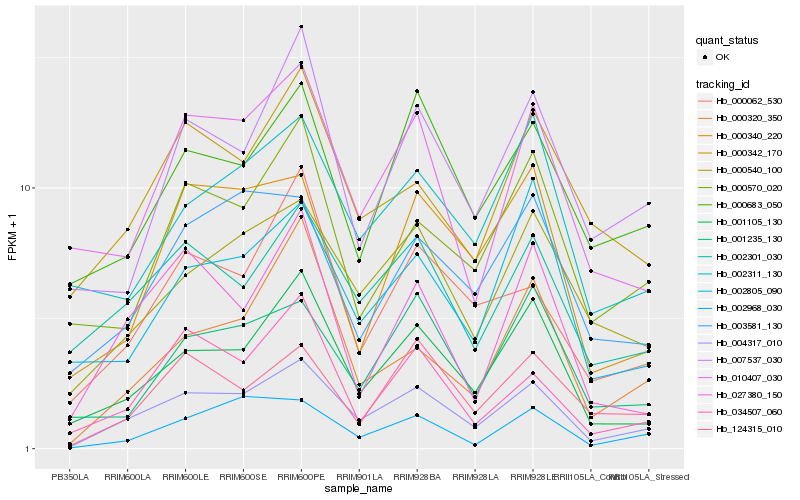
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004317_010 |
0.0 |
- |
- |
hypothetical protein CICLE_v10011038mg [Citrus clementina] |
| 2 |
Hb_001105_130 |
0.1177122282 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 3 |
Hb_000540_100 |
0.1328085963 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g60050 [Jatropha curcas] |
| 4 |
Hb_002301_030 |
0.1365958308 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1 [Jatropha curcas] |
| 5 |
Hb_000062_530 |
0.1439715575 |
- |
- |
PREDICTED: GDP-mannose transporter GONST3-like [Populus euphratica] |
| 6 |
Hb_010407_030 |
0.1482246197 |
- |
- |
PREDICTED: casein kinase I-like [Jatropha curcas] |
| 7 |
Hb_124315_010 |
0.1493821248 |
- |
- |
PREDICTED: uncharacterized protein LOC105650413 [Jatropha curcas] |
| 8 |
Hb_003581_130 |
0.152132175 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 9 |
Hb_034507_060 |
0.1527020698 |
- |
- |
PREDICTED: UPF0503 protein At3g09070, chloroplastic-like [Citrus sinensis] |
| 10 |
Hb_000320_350 |
0.152802998 |
- |
- |
hypothetical protein RCOM_0904330 [Ricinus communis] |
| 11 |
Hb_007537_030 |
0.1542352558 |
- |
- |
PREDICTED: gamma aminobutyrate transaminase 3, chloroplastic [Jatropha curcas] |
| 12 |
Hb_027380_150 |
0.1543704053 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002968_030 |
0.1546022242 |
- |
- |
Pectinesterase-1 precursor, putative [Ricinus communis] |
| 14 |
Hb_000342_170 |
0.1559753499 |
- |
- |
PREDICTED: uncharacterized protein LOC100807540 isoform X2 [Glycine max] |
| 15 |
Hb_000683_050 |
0.1561077281 |
- |
- |
PREDICTED: probable carbohydrate esterase At4g34215 [Jatropha curcas] |
| 16 |
Hb_001235_130 |
0.1563479166 |
- |
- |
- |
| 17 |
Hb_000340_220 |
0.1577962782 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000570_020 |
0.1578549325 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX9H isoform X1 [Jatropha curcas] |
| 19 |
Hb_002311_130 |
0.1587701362 |
- |
- |
PREDICTED: importin subunit alpha-2 isoform X2 [Jatropha curcas] |
| 20 |
Hb_002805_090 |
0.1592740544 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |