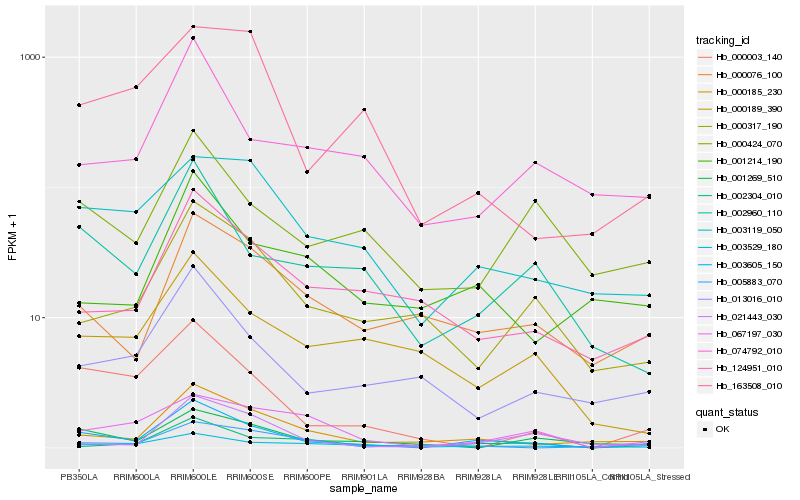
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003605_150 |
0.0 |
- |
- |
chitinase family protein [Populus trichocarpa] |
| 2 |
Hb_000185_230 |
0.2059831547 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 3 |
Hb_002960_110 |
0.2276127203 |
- |
- |
PREDICTED: synaptotagmin-2 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000003_140 |
0.2294333006 |
- |
- |
hypothetical protein glysoja_047034, partial [Glycine soja] |
| 5 |
Hb_003529_180 |
0.2405057741 |
- |
- |
hypothetical protein JCGZ_00222 [Jatropha curcas] |
| 6 |
Hb_003119_050 |
0.2426036382 |
- |
- |
PREDICTED: bifunctional riboflavin biosynthesis protein RIBA 1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_002304_010 |
0.2449382706 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 8 |
Hb_013016_010 |
0.2458524261 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000076_100 |
0.2488783129 |
- |
- |
PREDICTED: septum-promoting GTP-binding protein 1 [Jatropha curcas] |
| 10 |
Hb_074792_010 |
0.2524566748 |
- |
- |
PREDICTED: probable lactoylglutathione lyase, chloroplast [Gossypium raimondii] |
| 11 |
Hb_124951_010 |
0.2536200347 |
- |
- |
chitinase-like protein [Hevea brasiliensis] |
| 12 |
Hb_067197_030 |
0.2584952658 |
- |
- |
PREDICTED: UPF0187 protein At3g61320, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000424_070 |
0.2586858295 |
- |
- |
PREDICTED: serine/arginine repetitive matrix protein 2-like [Jatropha curcas] |
| 14 |
Hb_000317_190 |
0.2594739997 |
- |
- |
PREDICTED: diacylglycerol kinase 5-like [Jatropha curcas] |
| 15 |
Hb_021443_030 |
0.2600432401 |
- |
- |
PREDICTED: uncharacterized protein LOC105648566 [Jatropha curcas] |
| 16 |
Hb_163508_010 |
0.2611788355 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1 [Jatropha curcas] |
| 17 |
Hb_001214_190 |
0.2642602033 |
- |
- |
hypothetical protein JCGZ_25001 [Jatropha curcas] |
| 18 |
Hb_001269_510 |
0.2658617363 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 19 |
Hb_000189_390 |
0.2660424654 |
- |
- |
PREDICTED: pollen-specific protein SF21-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_005883_070 |
0.2678087788 |
- |
- |
PREDICTED: probable myosin-binding protein 5 [Jatropha curcas] |