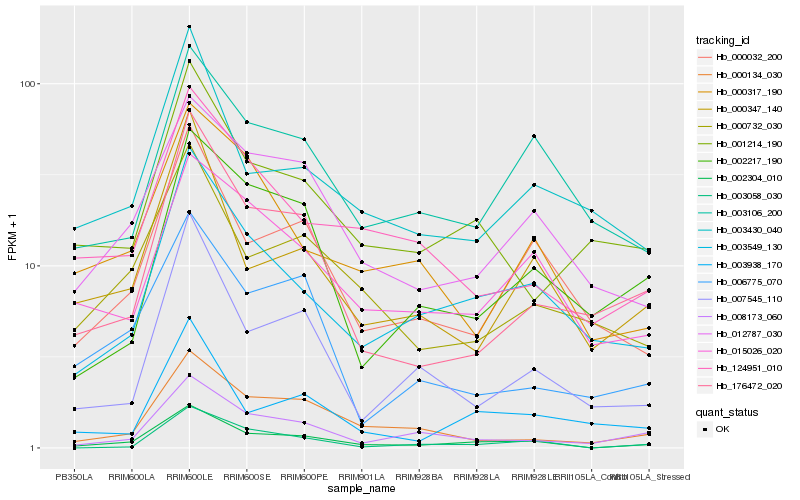
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002304_010 |
0.0 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 2 |
Hb_003549_130 |
0.157974956 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |
| 3 |
Hb_000032_200 |
0.1696356126 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 4 |
Hb_008173_060 |
0.1752505918 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 2 [Jatropha curcas] |
| 5 |
Hb_012787_030 |
0.175990057 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 1 [Jatropha curcas] |
| 6 |
Hb_003058_030 |
0.1787131607 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 7 |
Hb_176472_020 |
0.1793630799 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000347_140 |
0.1813545514 |
- |
- |
PREDICTED: protein FLUORESCENT IN BLUE LIGHT, chloroplastic isoform X2 [Jatropha curcas] |
| 9 |
Hb_007545_110 |
0.1814239186 |
- |
- |
catalytic, putative [Ricinus communis] |
| 10 |
Hb_000134_030 |
0.182109392 |
- |
- |
PREDICTED: sister chromatid cohesion 1 protein 3-like [Jatropha curcas] |
| 11 |
Hb_000732_030 |
0.1868775037 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_015026_020 |
0.1897390702 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os07g0563300 [Jatropha curcas] |
| 13 |
Hb_124951_010 |
0.1921096769 |
- |
- |
chitinase-like protein [Hevea brasiliensis] |
| 14 |
Hb_001214_190 |
0.1955408016 |
- |
- |
hypothetical protein JCGZ_25001 [Jatropha curcas] |
| 15 |
Hb_006775_070 |
0.1956846318 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 16 |
Hb_003430_040 |
0.1965231319 |
- |
- |
spermidine synthase 1, putative [Ricinus communis] |
| 17 |
Hb_003106_200 |
0.1965848564 |
- |
- |
PREDICTED: uncharacterized protein LOC105640430 isoform X1 [Jatropha curcas] |
| 18 |
Hb_003938_170 |
0.197322202 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase homolog 1-like [Jatropha curcas] |
| 19 |
Hb_002217_190 |
0.1980360926 |
- |
- |
invertase [Hevea brasiliensis] |
| 20 |
Hb_000317_190 |
0.1981193013 |
- |
- |
PREDICTED: diacylglycerol kinase 5-like [Jatropha curcas] |