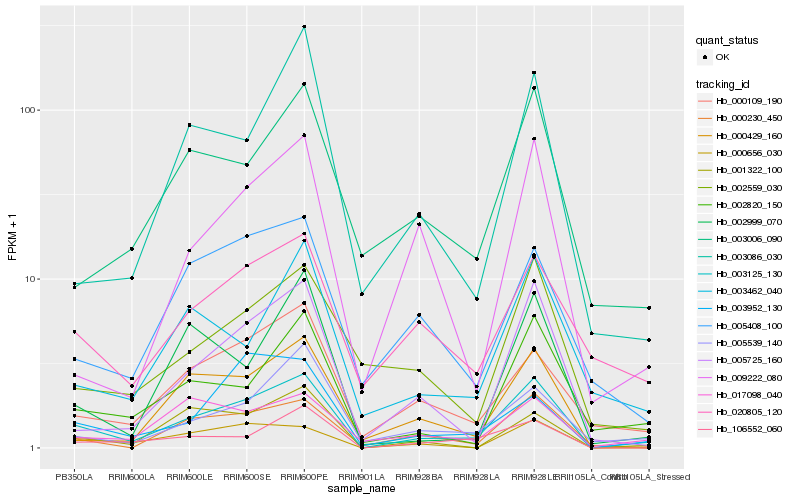
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003125_130 |
0.0 |
- |
- |
hypothetical protein JCGZ_26446 [Jatropha curcas] |
| 2 |
Hb_002820_150 |
0.1898624286 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 3 |
Hb_000230_450 |
0.1938716522 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_005725_160 |
0.1949953799 |
- |
- |
PREDICTED: uncharacterized protein LOC105630811 [Jatropha curcas] |
| 5 |
Hb_000429_160 |
0.1955428413 |
- |
- |
PREDICTED: uncharacterized protein LOC105645177 [Jatropha curcas] |
| 6 |
Hb_000109_190 |
0.1960476484 |
- |
- |
PREDICTED: RING-H2 finger protein ATL47 [Jatropha curcas] |
| 7 |
Hb_106552_060 |
0.1973323748 |
- |
- |
STS14 protein precursor, putative [Ricinus communis] |
| 8 |
Hb_000656_030 |
0.2050439678 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001322_100 |
0.2069728469 |
- |
- |
PREDICTED: hyoscyamine 6-dioxygenase [Vitis vinifera] |
| 10 |
Hb_005539_140 |
0.2077544984 |
- |
- |
PREDICTED: U-box domain-containing protein 19-like [Jatropha curcas] |
| 11 |
Hb_003006_090 |
0.2090419964 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 2 [UDP-forming] isoform X2 [Jatropha curcas] |
| 12 |
Hb_003462_040 |
0.2099431568 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 13 |
Hb_020805_120 |
0.2115991435 |
- |
- |
PREDICTED: glutamate receptor 3.3 [Jatropha curcas] |
| 14 |
Hb_017098_040 |
0.2138997125 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 15 |
Hb_002559_030 |
0.2152612298 |
- |
- |
oxidoreductase, putative [Ricinus communis] |
| 16 |
Hb_002999_070 |
0.2159259539 |
- |
- |
glucose-methanol-choline (gmc) oxidoreductase, putative [Ricinus communis] |
| 17 |
Hb_003086_030 |
0.2168864647 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 17 [Jatropha curcas] |
| 18 |
Hb_005408_100 |
0.218893262 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 14 [Jatropha curcas] |
| 19 |
Hb_003952_130 |
0.2191290685 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 20 [Jatropha curcas] |
| 20 |
Hb_009222_080 |
0.2216381169 |
- |
- |
RING-H2 finger protein ATL2J, putative [Ricinus communis] |