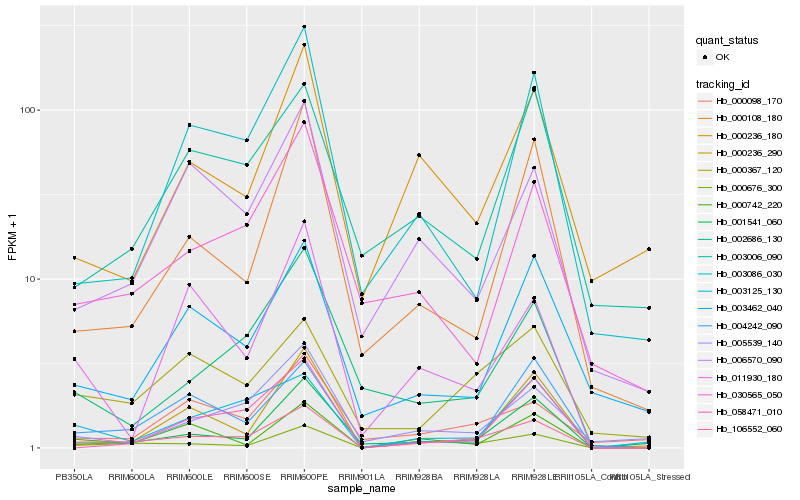
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_106552_060 |
0.0 |
- |
- |
STS14 protein precursor, putative [Ricinus communis] |
| 2 |
Hb_000676_300 |
0.1586586284 |
- |
- |
PREDICTED: probable caffeine synthase 4 [Jatropha curcas] |
| 3 |
Hb_003125_130 |
0.1973323748 |
- |
- |
hypothetical protein JCGZ_26446 [Jatropha curcas] |
| 4 |
Hb_000367_120 |
0.1985046895 |
transcription factor |
TF Family: C2H2 |
hypothetical protein RCOM_1470470 [Ricinus communis] |
| 5 |
Hb_001541_060 |
0.2015691822 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000742_220 |
0.2063034619 |
- |
- |
PREDICTED: uncharacterized protein LOC105633730 [Jatropha curcas] |
| 7 |
Hb_006570_090 |
0.2071528105 |
- |
- |
tubulin beta chain, putative [Ricinus communis] |
| 8 |
Hb_002686_130 |
0.2075102751 |
transcription factor |
TF Family: G2-like |
PREDICTED: uncharacterized protein LOC105650401 [Jatropha curcas] |
| 9 |
Hb_004242_090 |
0.2075473699 |
- |
- |
hypothetical protein JCGZ_00767 [Jatropha curcas] |
| 10 |
Hb_000108_180 |
0.2106929718 |
- |
- |
PREDICTED: uncharacterized protein LOC105630021 [Jatropha curcas] |
| 11 |
Hb_003086_030 |
0.2122468453 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 17 [Jatropha curcas] |
| 12 |
Hb_005539_140 |
0.2145095386 |
- |
- |
PREDICTED: U-box domain-containing protein 19-like [Jatropha curcas] |
| 13 |
Hb_000098_170 |
0.2147072688 |
- |
- |
hypothetical protein JCGZ_06491 [Jatropha curcas] |
| 14 |
Hb_003462_040 |
0.2168236341 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 15 |
Hb_030565_050 |
0.2169222761 |
- |
- |
UDP-D-GLUCURONATE 4-EPIMERASE 6 family protein [Populus trichocarpa] |
| 16 |
Hb_000236_290 |
0.2202644863 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 17 |
Hb_058471_010 |
0.2207938702 |
- |
- |
PREDICTED: clathrin light chain 1 [Jatropha curcas] |
| 18 |
Hb_003006_090 |
0.2231901731 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 2 [UDP-forming] isoform X2 [Jatropha curcas] |
| 19 |
Hb_011930_180 |
0.2254204684 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000236_180 |
0.2258472322 |
- |
- |
serine hydroxymethyltransferase, putative [Ricinus communis] |