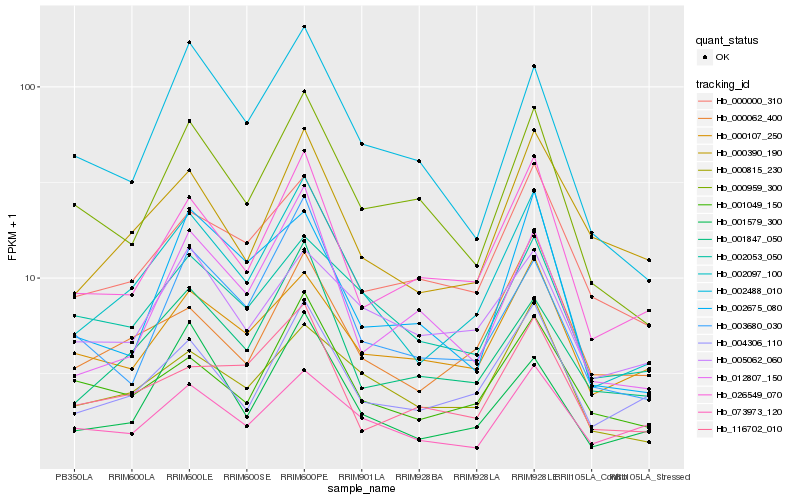
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002097_100 |
0.0 |
- |
- |
protein with unknown function [Ricinus communis] |
| 2 |
Hb_026549_070 |
0.1154244078 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 1 [Jatropha curcas] |
| 3 |
Hb_004306_110 |
0.1207966953 |
transcription factor |
TF Family: Trihelix |
PREDICTED: phosphatidylinositol/phosphatidylcholine transfer protein SFH9 [Jatropha curcas] |
| 4 |
Hb_000815_230 |
0.1252531647 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001847_050 |
0.1316832431 |
- |
- |
hypothetical protein CISIN_1g019843mg [Citrus sinensis] |
| 6 |
Hb_002053_050 |
0.1339701732 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 7 |
Hb_001579_300 |
0.135001955 |
- |
- |
PREDICTED: probable polygalacturonase At1g80170 [Jatropha curcas] |
| 8 |
Hb_001049_150 |
0.1357146497 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 11 [Jatropha curcas] |
| 9 |
Hb_000959_300 |
0.1367949057 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 2 [UDP-forming] isoform X2 [Jatropha curcas] |
| 10 |
Hb_000062_400 |
0.1393285869 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000000_310 |
0.1405719295 |
- |
- |
metalloendopeptidase, putative [Ricinus communis] |
| 12 |
Hb_002488_010 |
0.1434182619 |
- |
- |
PREDICTED: gamma-interferon-inducible-lysosomal thiol reductase [Jatropha curcas] |
| 13 |
Hb_000107_250 |
0.1441440796 |
- |
- |
voltage-gated clc-type chloride channel, putative [Ricinus communis] |
| 14 |
Hb_000390_190 |
0.1446620244 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 15 |
Hb_003680_030 |
0.1458671626 |
- |
- |
PREDICTED: aspartic proteinase-like protein 2 isoform X3 [Jatropha curcas] |
| 16 |
Hb_012807_150 |
0.1469735923 |
- |
- |
PREDICTED: alpha-L-fucosidase 1 [Jatropha curcas] |
| 17 |
Hb_073973_120 |
0.1469780467 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 18 |
Hb_116702_010 |
0.1480322982 |
- |
- |
Leucine-rich repeat protein kinase family protein [Theobroma cacao] |
| 19 |
Hb_005062_060 |
0.1480608093 |
- |
- |
PREDICTED: uncharacterized protein At4g06598-like [Jatropha curcas] |
| 20 |
Hb_002675_080 |
0.1497615781 |
- |
- |
PREDICTED: uncharacterized protein LOC105634950 [Jatropha curcas] |