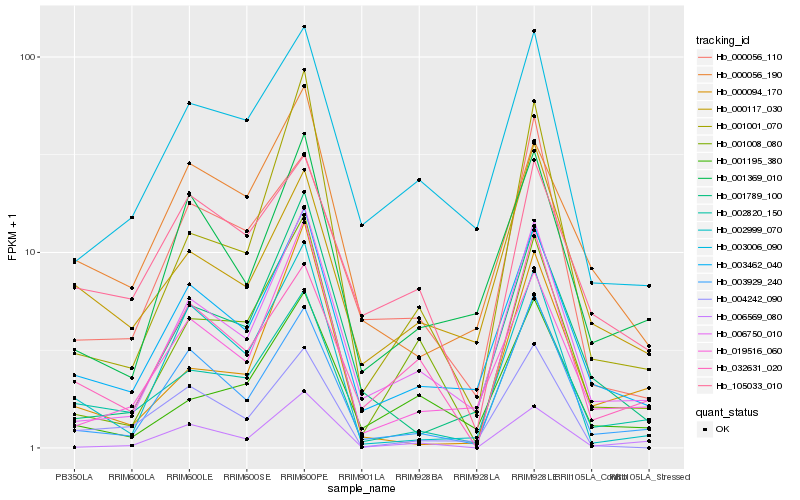
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002820_150 |
0.0 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 2 |
Hb_003462_040 |
0.1211664469 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 3 |
Hb_003929_240 |
0.1412335398 |
- |
- |
PREDICTED: uncharacterized protein LOC105643616 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000117_030 |
0.149258629 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 [Jatropha curcas] |
| 5 |
Hb_006750_010 |
0.1570362212 |
- |
- |
Beta-1,3-galactosyltransferase sqv-2, putative [Ricinus communis] |
| 6 |
Hb_000094_170 |
0.1611953201 |
- |
- |
PREDICTED: uncharacterized protein LOC105633781 [Jatropha curcas] |
| 7 |
Hb_004242_090 |
0.1628594542 |
- |
- |
hypothetical protein JCGZ_00767 [Jatropha curcas] |
| 8 |
Hb_019516_060 |
0.1681827302 |
- |
- |
PREDICTED: uncharacterized protein LOC105634259 [Jatropha curcas] |
| 9 |
Hb_000056_190 |
0.1683972367 |
- |
- |
hypothetical protein [Ricinus communis] |
| 10 |
Hb_001008_080 |
0.172633775 |
- |
- |
PREDICTED: acyl-protein thioesterase 2 [Jatropha curcas] |
| 11 |
Hb_000056_110 |
0.1728234945 |
- |
- |
PREDICTED: uncharacterized protein LOC105630405 [Jatropha curcas] |
| 12 |
Hb_001001_070 |
0.1734866212 |
- |
- |
PREDICTED: uncharacterized protein LOC105633663 [Jatropha curcas] |
| 13 |
Hb_032631_020 |
0.1737228255 |
- |
- |
glycine-rich family protein [Populus trichocarpa] |
| 14 |
Hb_002999_070 |
0.1739993171 |
- |
- |
glucose-methanol-choline (gmc) oxidoreductase, putative [Ricinus communis] |
| 15 |
Hb_001789_100 |
0.1744104526 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001195_380 |
0.1745155942 |
- |
- |
PREDICTED: uncharacterized protein LOC105633781 [Jatropha curcas] |
| 17 |
Hb_003006_090 |
0.1749145537 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 2 [UDP-forming] isoform X2 [Jatropha curcas] |
| 18 |
Hb_001369_010 |
0.1781553473 |
- |
- |
Nodulation receptor kinase precursor, putative [Ricinus communis] |
| 19 |
Hb_105033_010 |
0.1812411527 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 2 isoform X2 [Jatropha curcas] |
| 20 |
Hb_006569_080 |
0.1818226747 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein VITISV_024559 [Vitis vinifera] |