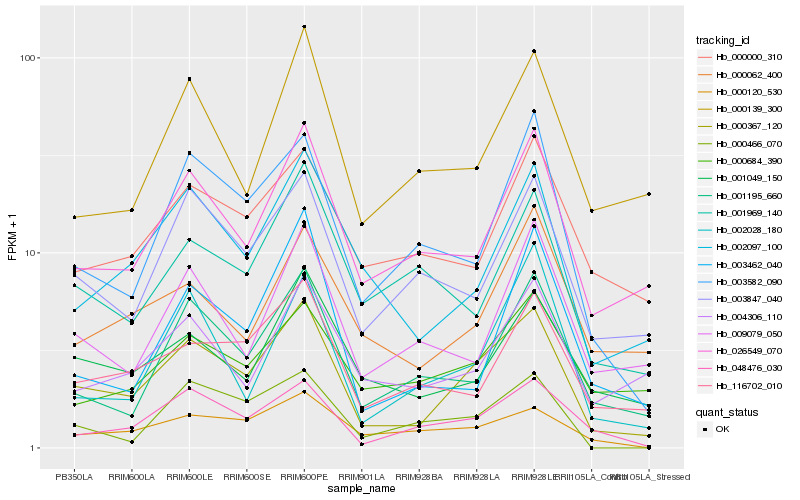
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000367_120 |
0.0 |
transcription factor |
TF Family: C2H2 |
hypothetical protein RCOM_1470470 [Ricinus communis] |
| 2 |
Hb_002097_100 |
0.1554429945 |
- |
- |
protein with unknown function [Ricinus communis] |
| 3 |
Hb_026549_070 |
0.1556514892 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 1 [Jatropha curcas] |
| 4 |
Hb_048476_030 |
0.1557298371 |
- |
- |
hypothetical protein POPTR_0010s11560g [Populus trichocarpa] |
| 5 |
Hb_003582_090 |
0.1634885105 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 6-like [Jatropha curcas] |
| 6 |
Hb_001195_660 |
0.1637251888 |
- |
- |
metalloendopeptidase, putative [Ricinus communis] |
| 7 |
Hb_116702_010 |
0.1667672838 |
- |
- |
Leucine-rich repeat protein kinase family protein [Theobroma cacao] |
| 8 |
Hb_003847_040 |
0.1671504046 |
- |
- |
adenosine diphosphatase, putative [Ricinus communis] |
| 9 |
Hb_001049_150 |
0.169375036 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 11 [Jatropha curcas] |
| 10 |
Hb_000120_530 |
0.175349241 |
transcription factor |
TF Family: C3H |
RING finger protein 113A, putative [Ricinus communis] |
| 11 |
Hb_000139_300 |
0.1832145446 |
- |
- |
hypothetical protein POPTR_0008s19950g [Populus trichocarpa] |
| 12 |
Hb_002028_180 |
0.1836446449 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 13 |
Hb_000062_400 |
0.1840424478 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_009079_050 |
0.1855417711 |
- |
- |
PREDICTED: V-type proton ATPase subunit a3-like [Jatropha curcas] |
| 15 |
Hb_004306_110 |
0.1860725456 |
transcription factor |
TF Family: Trihelix |
PREDICTED: phosphatidylinositol/phosphatidylcholine transfer protein SFH9 [Jatropha curcas] |
| 16 |
Hb_001969_140 |
0.1877386178 |
- |
- |
PREDICTED: uncharacterized protein LOC105634929 [Jatropha curcas] |
| 17 |
Hb_000684_390 |
0.1884546117 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 4 [Jatropha curcas] |
| 18 |
Hb_000466_070 |
0.188788551 |
- |
- |
hypothetical protein JCGZ_08989 [Jatropha curcas] |
| 19 |
Hb_003462_040 |
0.1888538287 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 20 |
Hb_000000_310 |
0.1890666549 |
- |
- |
metalloendopeptidase, putative [Ricinus communis] |