| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
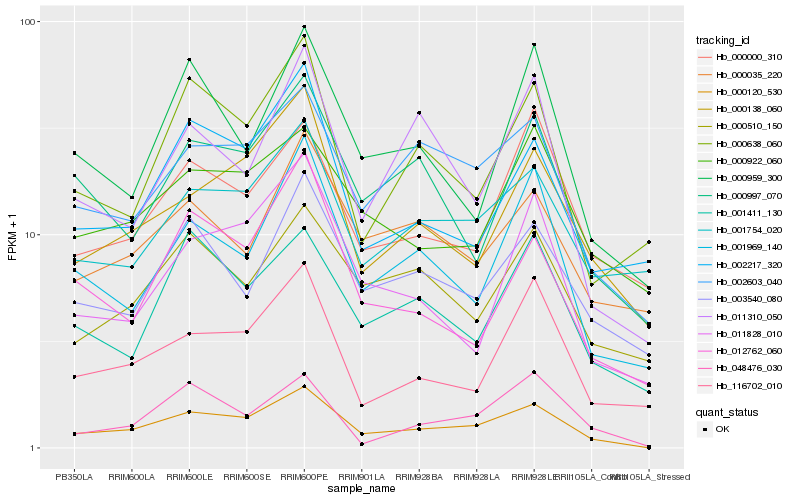
Hb_000120_530 |
0.0 |
transcription factor |
TF Family: C3H |
RING finger protein 113A, putative [Ricinus communis] |
| 2 |
Hb_002603_040 |
0.1269981759 |
- |
- |
PREDICTED: phosphoinositide phospholipase C 2-like [Jatropha curcas] |
| 3 |
Hb_000138_060 |
0.1462430188 |
- |
- |
PREDICTED: protein trichome birefringence [Jatropha curcas] |
| 4 |
Hb_001969_140 |
0.1487588371 |
- |
- |
PREDICTED: uncharacterized protein LOC105634929 [Jatropha curcas] |
| 5 |
Hb_000997_070 |
0.1544349268 |
- |
- |
PREDICTED: ureidoglycolate hydrolase isoform X1 [Jatropha curcas] |
| 6 |
Hb_003540_080 |
0.1553848611 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 7 |
Hb_001411_130 |
0.1572610126 |
- |
- |
PREDICTED: uncharacterized protein LOC105631354 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001754_020 |
0.1578458573 |
- |
- |
PREDICTED: kinesin-13A isoform X1 [Jatropha curcas] |
| 9 |
Hb_011828_010 |
0.1581167878 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g67720 [Jatropha curcas] |
| 10 |
Hb_116702_010 |
0.1582135967 |
- |
- |
Leucine-rich repeat protein kinase family protein [Theobroma cacao] |
| 11 |
Hb_000638_060 |
0.1591136042 |
- |
- |
PREDICTED: probable methyltransferase PMT26 [Jatropha curcas] |
| 12 |
Hb_000922_060 |
0.1599005891 |
- |
- |
kinesin light chain, putative [Ricinus communis] |
| 13 |
Hb_011310_050 |
0.1600631189 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 1 [UDP-forming] [Jatropha curcas] |
| 14 |
Hb_048476_030 |
0.161729245 |
- |
- |
hypothetical protein POPTR_0010s11560g [Populus trichocarpa] |
| 15 |
Hb_000959_300 |
0.1623497415 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 2 [UDP-forming] isoform X2 [Jatropha curcas] |
| 16 |
Hb_002217_320 |
0.1624138057 |
- |
- |
PREDICTED: probable O-acetyltransferase CAS1 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000510_150 |
0.1624426388 |
- |
- |
PREDICTED: magnesium transporter MRS2-I-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_000000_310 |
0.1631878238 |
- |
- |
metalloendopeptidase, putative [Ricinus communis] |
| 19 |
Hb_012762_060 |
0.1647302001 |
- |
- |
Vitellogenic carboxypeptidase, putative [Ricinus communis] |
| 20 |
Hb_000035_220 |
0.1648556102 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 8 [Jatropha curcas] |