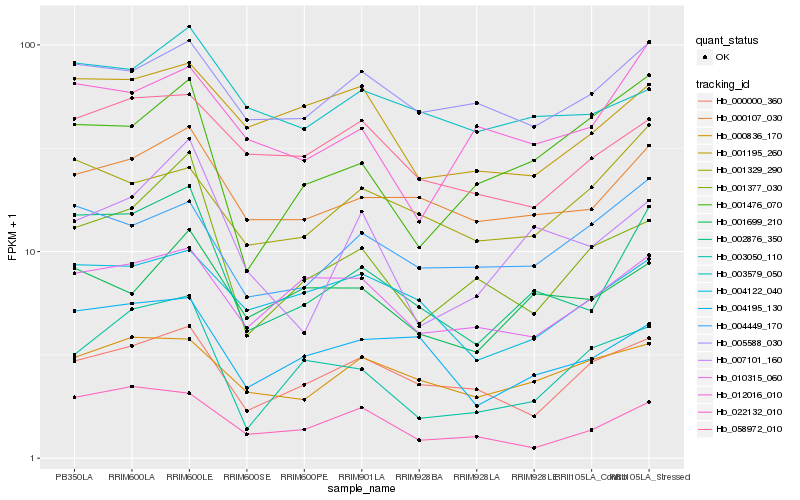
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002876_350 |
0.0 |
- |
- |
PREDICTED: gibberellin-regulated protein 9-like isoform X2 [Citrus sinensis] |
| 2 |
Hb_004195_130 |
0.1233701182 |
- |
- |
PREDICTED: thioredoxin domain-containing protein PLP3B [Sesamum indicum] |
| 3 |
Hb_000107_030 |
0.1345099178 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_022132_010 |
0.1363021157 |
- |
- |
unknown [Medicago truncatula] |
| 5 |
Hb_001377_030 |
0.1413886472 |
- |
- |
PREDICTED: histone H3-like centromeric protein HTR12 [Jatropha curcas] |
| 6 |
Hb_007101_160 |
0.1504390902 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_004122_040 |
0.15577266 |
- |
- |
PREDICTED: polyadenylate-binding protein 2-like [Jatropha curcas] |
| 8 |
Hb_001699_210 |
0.1557991981 |
- |
- |
PREDICTED: DNA-(apurinic or apyrimidinic site) lyase 2 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000000_360 |
0.1571602624 |
- |
- |
PREDICTED: methyltransferase-like protein 2 [Populus euphratica] |
| 10 |
Hb_010315_060 |
0.1573334189 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 [Jatropha curcas] |
| 11 |
Hb_004449_170 |
0.1575324386 |
- |
- |
PREDICTED: uncharacterized protein LOC105631285 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000836_170 |
0.1581534906 |
- |
- |
PREDICTED: RAB6A-GEF complex partner protein 1-like [Jatropha curcas] |
| 13 |
Hb_058972_010 |
0.1581834023 |
- |
- |
PREDICTED: uncharacterized protein LOC105644404 isoform X2 [Jatropha curcas] |
| 14 |
Hb_003050_110 |
0.1585823153 |
- |
- |
PREDICTED: prefoldin subunit 6 [Populus euphratica] |
| 15 |
Hb_003579_050 |
0.1587282955 |
- |
- |
PREDICTED: 14-3-3 protein 7 [Jatropha curcas] |
| 16 |
Hb_001195_260 |
0.1593305851 |
- |
- |
Mitochondrial 50S ribosomal protein L27 [Hevea brasiliensis] |
| 17 |
Hb_012016_010 |
0.1603341039 |
- |
- |
PREDICTED: glycine-rich RNA-binding protein 6, mitochondrial [Jatropha curcas] |
| 18 |
Hb_001329_290 |
0.1628405905 |
- |
- |
PREDICTED: uncharacterized protein LOC105642699 [Jatropha curcas] |
| 19 |
Hb_005588_030 |
0.163488643 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 14 homolog [Eucalyptus grandis] |
| 20 |
Hb_001476_070 |
0.1638762756 |
- |
- |
MinE protein [Manihot esculenta] |