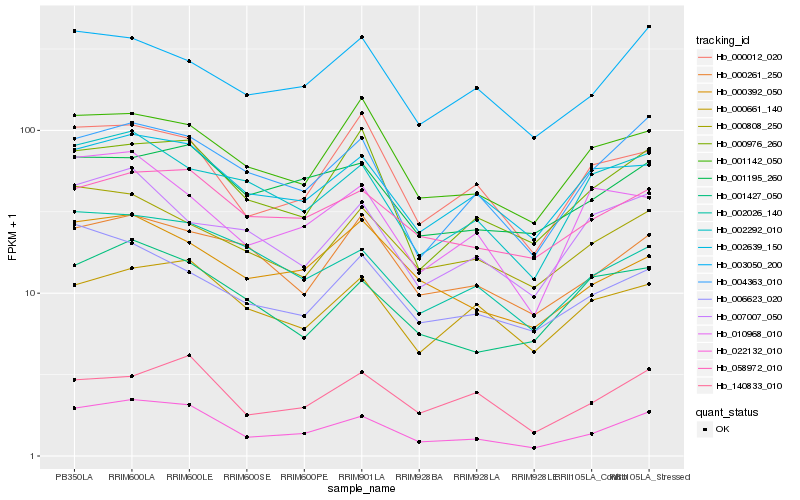
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_022132_010 |
0.0 |
- |
- |
unknown [Medicago truncatula] |
| 2 |
Hb_004363_010 |
0.1042758669 |
- |
- |
hypothetical protein CISIN_1g0405721mg, partial [Citrus sinensis] |
| 3 |
Hb_000976_260 |
0.1094086741 |
- |
- |
- |
| 4 |
Hb_002026_140 |
0.1119629594 |
- |
- |
hypothetical protein POPTR_0006s13340g [Populus trichocarpa] |
| 5 |
Hb_002639_150 |
0.1133439143 |
- |
- |
- |
| 6 |
Hb_002292_010 |
0.1156574025 |
- |
- |
hypothetical protein CICLE_v10029628mg [Citrus clementina] |
| 7 |
Hb_000012_020 |
0.116401859 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 20 [Jatropha curcas] |
| 8 |
Hb_058972_010 |
0.1175840247 |
- |
- |
PREDICTED: uncharacterized protein LOC105644404 isoform X2 [Jatropha curcas] |
| 9 |
Hb_140833_010 |
0.1196665592 |
- |
- |
PREDICTED: uncharacterized protein LOC105647767 [Jatropha curcas] |
| 10 |
Hb_001195_260 |
0.1205012031 |
- |
- |
Mitochondrial 50S ribosomal protein L27 [Hevea brasiliensis] |
| 11 |
Hb_001427_050 |
0.1208942814 |
- |
- |
PREDICTED: probable protein arginine N-methyltransferase 1.2 [Jatropha curcas] |
| 12 |
Hb_000392_050 |
0.1216833175 |
- |
- |
PREDICTED: uncharacterized protein LOC105640540 isoform X2 [Jatropha curcas] |
| 13 |
Hb_001142_050 |
0.12215654 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 12 [Jatropha curcas] |
| 14 |
Hb_003050_200 |
0.1228884174 |
- |
- |
Ribosomal protein L18ae/LX family protein [Theobroma cacao] |
| 15 |
Hb_000808_250 |
0.123245766 |
- |
- |
PREDICTED: uncharacterized protein LOC105647419 [Jatropha curcas] |
| 16 |
Hb_010968_010 |
0.1266318476 |
- |
- |
Golgi snare 12 isoform 1 [Theobroma cacao] |
| 17 |
Hb_006623_020 |
0.1274280657 |
- |
- |
PREDICTED: protein LURP-one-related 7 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000261_250 |
0.1281534059 |
- |
- |
elongation factor 1-alpha [Lepidium sativum] |
| 19 |
Hb_000661_140 |
0.1284853152 |
- |
- |
PREDICTED: uncharacterized protein LOC105648317 isoform X2 [Jatropha curcas] |
| 20 |
Hb_007007_050 |
0.1295236815 |
- |
- |
PREDICTED: ubiquitin-related modifier 1 homolog 2 [Malus domestica] |