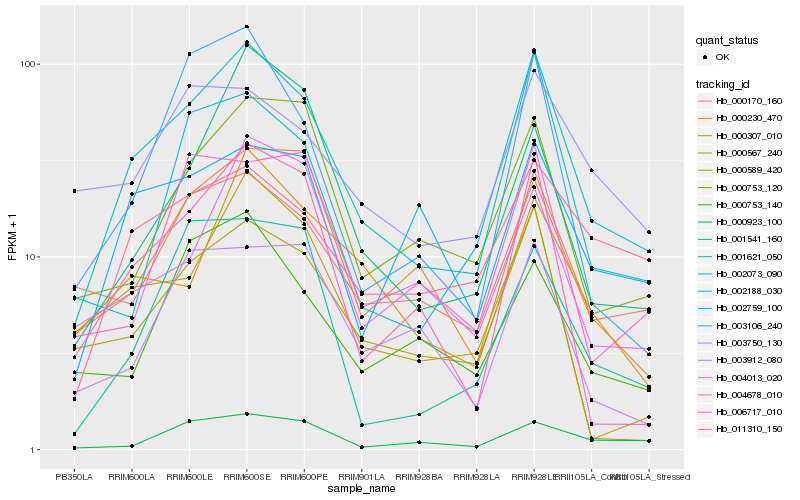
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002073_090 |
0.0 |
- |
- |
PREDICTED: copper transporter 6-like [Jatropha curcas] |
| 2 |
Hb_006717_010 |
0.1654406592 |
- |
- |
PREDICTED: uncharacterized protein LOC105649338 [Jatropha curcas] |
| 3 |
Hb_000230_470 |
0.1692155238 |
- |
- |
PREDICTED: mitogen-activated protein kinase 15 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000170_160 |
0.1693043822 |
- |
- |
PREDICTED: NAD kinase 2, chloroplastic [Jatropha curcas] |
| 5 |
Hb_003106_240 |
0.1738050801 |
- |
- |
hypothetical protein POPTR_0012s07880g [Populus trichocarpa] |
| 6 |
Hb_000753_120 |
0.1757944288 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 7 |
Hb_011310_150 |
0.178536343 |
- |
- |
PREDICTED: SPX domain-containing protein 1-like [Jatropha curcas] |
| 8 |
Hb_000307_010 |
0.1789541866 |
- |
- |
PREDICTED: ganglioside-induced differentiation-associated protein 2-like [Jatropha curcas] |
| 9 |
Hb_000567_240 |
0.1795877666 |
- |
- |
PREDICTED: F-box/LRR-repeat MAX2 homolog A-like [Jatropha curcas] |
| 10 |
Hb_000589_420 |
0.18239767 |
- |
- |
PREDICTED: uncharacterized protein LOC105647273 [Jatropha curcas] |
| 11 |
Hb_000923_100 |
0.1852775868 |
- |
- |
chloride channel clc, putative [Ricinus communis] |
| 12 |
Hb_002759_100 |
0.1870560795 |
- |
- |
hypothetical protein JCGZ_21777 [Jatropha curcas] |
| 13 |
Hb_002188_030 |
0.1871643273 |
- |
- |
PREDICTED: probable zinc metallopeptidase EGY3, chloroplastic isoform X1 [Jatropha curcas] |
| 14 |
Hb_003750_130 |
0.1880954331 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001621_050 |
0.189676978 |
- |
- |
PREDICTED: uncharacterized protein LOC105647273 [Jatropha curcas] |
| 16 |
Hb_004678_010 |
0.1898337429 |
transcription factor |
TF Family: Orphans |
Salt-tolerance protein, putative [Ricinus communis] |
| 17 |
Hb_004013_020 |
0.190043024 |
- |
- |
PREDICTED: F-box protein SKIP2 [Jatropha curcas] |
| 18 |
Hb_000753_140 |
0.1901548333 |
- |
- |
PREDICTED: uncharacterized protein LOC105640672 [Jatropha curcas] |
| 19 |
Hb_003912_080 |
0.1923047924 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 20 |
Hb_001541_160 |
0.1928302284 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase XERICO [Jatropha curcas] |