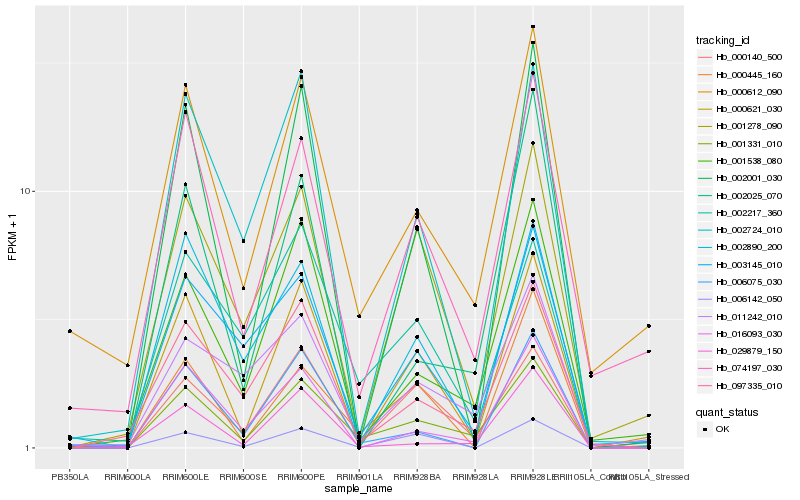
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001331_010 |
0.0 |
- |
- |
hypothetical protein CISIN_1g039303mg, partial [Citrus sinensis] |
| 2 |
Hb_001538_080 |
0.1578870585 |
- |
- |
PREDICTED: QWRF motif-containing protein 2 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000140_500 |
0.1592148662 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 4 |
Hb_000621_030 |
0.1607485327 |
- |
- |
PREDICTED: uncharacterized protein LOC105638736 isoform X1 [Jatropha curcas] |
| 5 |
Hb_097335_010 |
0.1611726927 |
- |
- |
abhydrolase domain containing, putative [Ricinus communis] |
| 6 |
Hb_016093_030 |
0.1680039918 |
- |
- |
ankyrin repeat protein [Bruguiera gymnorhiza] |
| 7 |
Hb_000612_090 |
0.1685647426 |
- |
- |
PREDICTED: mitochondrial carnitine/acylcarnitine carrier-like protein [Jatropha curcas] |
| 8 |
Hb_074197_030 |
0.174797804 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 9 |
Hb_002001_030 |
0.1768843907 |
- |
- |
Plasma membrane ATPase 4 [Gossypium arboreum] |
| 10 |
Hb_002217_360 |
0.1774154008 |
- |
- |
PREDICTED: nuclear poly(A) polymerase 3 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001278_090 |
0.1786762007 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 12 |
Hb_000445_160 |
0.1815506866 |
- |
- |
PREDICTED: probable calcium-binding protein CML44 [Jatropha curcas] |
| 13 |
Hb_006142_050 |
0.1816366879 |
- |
- |
LRR receptor-like serine/threonine-protein kinase, putative [Theobroma cacao] |
| 14 |
Hb_003145_010 |
0.1817975843 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At1g59780 [Jatropha curcas] |
| 15 |
Hb_006075_030 |
0.1822554761 |
- |
- |
PREDICTED: uncharacterized protein LOC105641890 isoform X1 [Jatropha curcas] |
| 16 |
Hb_002724_010 |
0.1827847424 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 4-like [Citrus sinensis] |
| 17 |
Hb_011242_010 |
0.1830415173 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-2 [Jatropha curcas] |
| 18 |
Hb_029879_150 |
0.186033583 |
desease resistance |
Gene Name: NB-ARC |
putative disease resistance gene NBS-LRR family protein [Populus trichocarpa] |
| 19 |
Hb_002025_070 |
0.1889418278 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g13770, chloroplastic-like isoform X1 [Populus euphratica] |
| 20 |
Hb_002890_200 |
0.1895375787 |
- |
- |
PREDICTED: methionine S-methyltransferase [Jatropha curcas] |