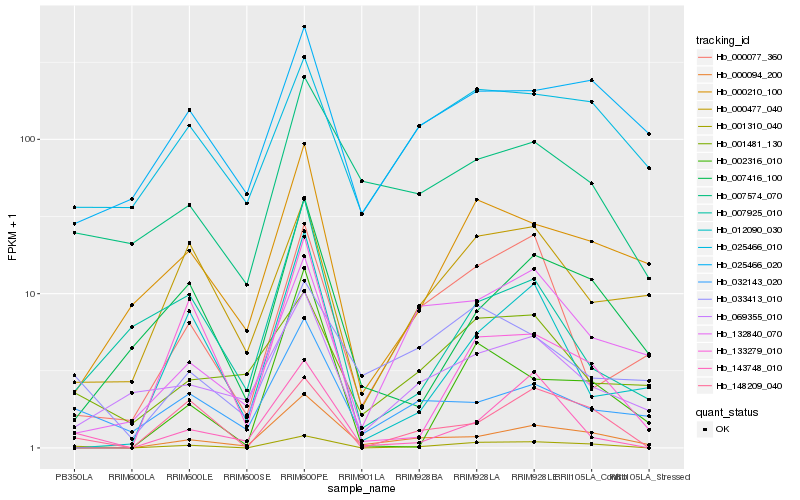
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001310_040 |
0.0 |
transcription factor |
TF Family: TAZ |
transcription cofactor, putative [Ricinus communis] |
| 2 |
Hb_148209_040 |
0.185957814 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_143748_010 |
0.2619846873 |
- |
- |
PREDICTED: uncharacterized protein LOC105648045 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000094_200 |
0.2699827087 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000477_040 |
0.2829568174 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000210_100 |
0.2832029348 |
- |
- |
pepsin A, putative [Ricinus communis] |
| 7 |
Hb_025466_010 |
0.289526676 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 8 |
Hb_007574_070 |
0.2900847096 |
- |
- |
fatty acid desaturase, putative [Ricinus communis] |
| 9 |
Hb_007416_100 |
0.2902166046 |
- |
- |
PREDICTED: probable xyloglucan glycosyltransferase 5 [Jatropha curcas] |
| 10 |
Hb_069355_010 |
0.2906728932 |
- |
- |
NBS-LRR resistance protein RGH2 [Manihot esculenta] |
| 11 |
Hb_032143_020 |
0.2913738284 |
- |
- |
hypothetical protein POPTR_0009s06310g [Populus trichocarpa] |
| 12 |
Hb_025466_020 |
0.2921821475 |
- |
- |
tubulin beta-6 [Brassica oleracea var. italica] |
| 13 |
Hb_000077_360 |
0.2941892157 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 14 |
Hb_132840_070 |
0.2984664484 |
- |
- |
metal tolerance protein 1 [Populus trichocarpa x Populus deltoides] |
| 15 |
Hb_033413_010 |
0.2991384118 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 16 |
Hb_001481_130 |
0.2992499062 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_133279_010 |
0.300673217 |
- |
- |
spermidine synthase 1, putative [Ricinus communis] |
| 18 |
Hb_012090_030 |
0.3023415914 |
- |
- |
PREDICTED: autophagy-related protein 8C isoform X2 [Phoenix dactylifera] |
| 19 |
Hb_002316_010 |
0.304714836 |
- |
- |
hypothetical protein JCGZ_11498 [Jatropha curcas] |
| 20 |
Hb_007925_010 |
0.3057163529 |
- |
- |
PREDICTED: asparagine synthetase [glutamine-hydrolyzing] 1 [Jatropha curcas] |