| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
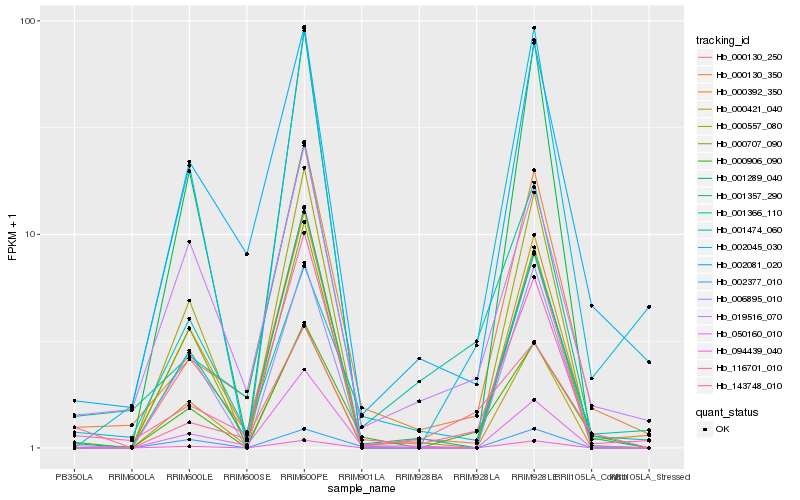
Hb_000906_090 |
0.0 |
- |
- |
hypothetical protein B456_002G267300 [Gossypium raimondii] |
| 2 |
Hb_000130_350 |
0.1849085448 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 3 |
Hb_116701_010 |
0.1901092677 |
- |
- |
hypothetical protein POPTR_0005s19410g [Populus trichocarpa] |
| 4 |
Hb_143748_010 |
0.1916308945 |
- |
- |
PREDICTED: uncharacterized protein LOC105648045 isoform X1 [Jatropha curcas] |
| 5 |
Hb_019516_070 |
0.19181877 |
- |
- |
PREDICTED: uncharacterized protein LOC105634258 [Jatropha curcas] |
| 6 |
Hb_001357_290 |
0.192796706 |
- |
- |
PREDICTED: uncharacterized protein LOC105632986 [Jatropha curcas] |
| 7 |
Hb_002045_030 |
0.1966355553 |
transcription factor |
TF Family: AUX/IAA |
hypothetical protein CISIN_1g047687mg [Citrus sinensis] |
| 8 |
Hb_006895_010 |
0.2066358942 |
- |
- |
PREDICTED: secologanin synthase-like [Citrus sinensis] |
| 9 |
Hb_002081_020 |
0.2071677673 |
transcription factor |
TF Family: LIM |
Cysteine and glycine-rich protein, putative [Ricinus communis] |
| 10 |
Hb_000392_350 |
0.2074098109 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 11 |
Hb_000421_040 |
0.2084016143 |
- |
- |
Peptide transporter, putative [Ricinus communis] |
| 12 |
Hb_000707_090 |
0.2102602616 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.6-like [Jatropha curcas] |
| 13 |
Hb_000557_080 |
0.2117634 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_050160_010 |
0.2135839083 |
- |
- |
hypothetical protein JCGZ_24934 [Jatropha curcas] |
| 15 |
Hb_001366_110 |
0.2147175551 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002377_010 |
0.2147234678 |
transcription factor |
TF Family: C2C2-CO-like |
PREDICTED: zinc finger protein CONSTANS-LIKE 2-like [Jatropha curcas] |
| 17 |
Hb_001474_060 |
0.2155294483 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 16 [Jatropha curcas] |
| 18 |
Hb_001289_040 |
0.2155956827 |
- |
- |
NDR1/HIN1-like 1 [Theobroma cacao] |
| 19 |
Hb_094439_040 |
0.2167303822 |
- |
- |
PREDICTED: cytochrome P450 93A1-like [Citrus sinensis] |
| 20 |
Hb_000130_250 |
0.2169490681 |
- |
- |
PREDICTED: protein trichome birefringence-like 31 [Jatropha curcas] |