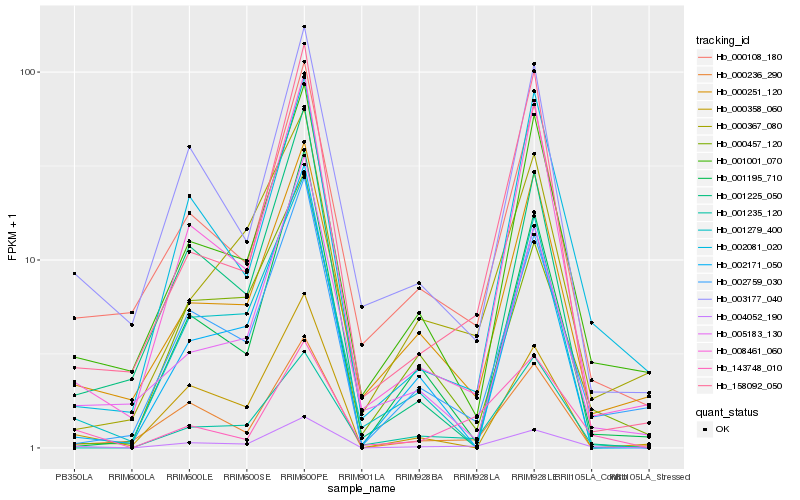
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004052_190 |
0.0 |
- |
- |
PREDICTED: COBRA-like protein 4 [Jatropha curcas] |
| 2 |
Hb_000108_180 |
0.1480587155 |
- |
- |
PREDICTED: uncharacterized protein LOC105630021 [Jatropha curcas] |
| 3 |
Hb_008461_060 |
0.1500795361 |
- |
- |
hypothetical protein JCGZ_17379 [Jatropha curcas] |
| 4 |
Hb_002759_030 |
0.1503119279 |
- |
- |
PREDICTED: serine/threonine-protein kinase PBS1 [Jatropha curcas] |
| 5 |
Hb_001001_070 |
0.1528483298 |
- |
- |
PREDICTED: uncharacterized protein LOC105633663 [Jatropha curcas] |
| 6 |
Hb_000251_120 |
0.1584030769 |
- |
- |
O-acetyltransferase, putative [Ricinus communis] |
| 7 |
Hb_003177_040 |
0.160364644 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 8 |
Hb_000236_290 |
0.1672853492 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 9 |
Hb_002081_020 |
0.1680465467 |
transcription factor |
TF Family: LIM |
Cysteine and glycine-rich protein, putative [Ricinus communis] |
| 10 |
Hb_001235_120 |
0.1685234227 |
- |
- |
hypothetical protein JCGZ_03395 [Jatropha curcas] |
| 11 |
Hb_001279_400 |
0.1690999539 |
- |
- |
PREDICTED: uncharacterized protein LOC105633544 [Jatropha curcas] |
| 12 |
Hb_158092_050 |
0.1694408696 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX10 [Jatropha curcas] |
| 13 |
Hb_143748_010 |
0.1696126323 |
- |
- |
PREDICTED: uncharacterized protein LOC105648045 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000358_060 |
0.1701014138 |
- |
- |
PREDICTED: pathogenesis-related protein 5-like [Jatropha curcas] |
| 15 |
Hb_000367_080 |
0.1735164693 |
- |
- |
PREDICTED: UDP-glucose 6-dehydrogenase 1-like [Jatropha curcas] |
| 16 |
Hb_001195_710 |
0.1752362753 |
- |
- |
glycosyltransferase family GT8 protein [Populus deltoides] |
| 17 |
Hb_000457_120 |
0.1768240725 |
- |
- |
PREDICTED: death-associated protein kinase dapk-1 [Jatropha curcas] |
| 18 |
Hb_005183_130 |
0.1794648152 |
- |
- |
glycogenin, putative [Ricinus communis] |
| 19 |
Hb_001225_050 |
0.1797380573 |
transcription factor |
TF Family: LIM |
Cysteine and glycine-rich protein, putative [Ricinus communis] |
| 20 |
Hb_002171_050 |
0.1797509941 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like protein kinase PEPR1 [Populus euphratica] |