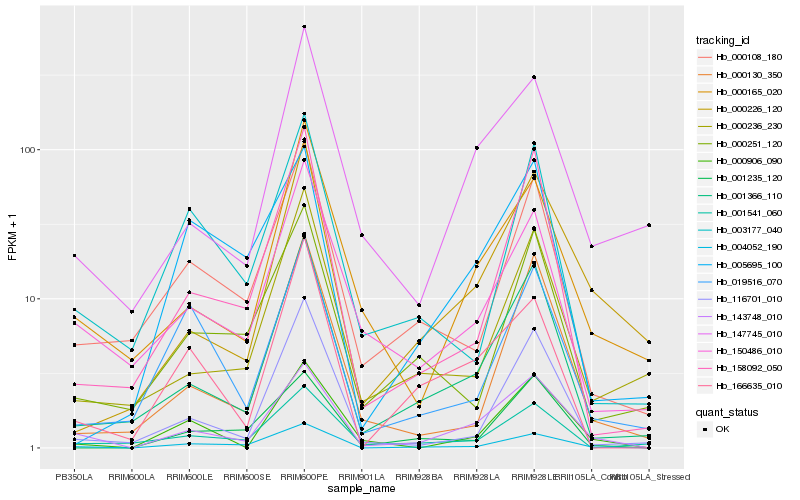
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_143748_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105648045 isoform X1 [Jatropha curcas] |
| 2 |
Hb_004052_190 |
0.1696126323 |
- |
- |
PREDICTED: COBRA-like protein 4 [Jatropha curcas] |
| 3 |
Hb_001366_110 |
0.1763393695 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000906_090 |
0.1916308945 |
- |
- |
hypothetical protein B456_002G267300 [Gossypium raimondii] |
| 5 |
Hb_147745_010 |
0.1926905691 |
- |
- |
PREDICTED: uncharacterized protein LOC105639149 [Jatropha curcas] |
| 6 |
Hb_000165_020 |
0.1945251339 |
- |
- |
PREDICTED: protein trichome birefringence-like 33 [Jatropha curcas] |
| 7 |
Hb_000226_120 |
0.1964967681 |
- |
- |
PREDICTED: heparanase-like protein 3 isoform X1 [Jatropha curcas] |
| 8 |
Hb_150486_010 |
0.1985176442 |
- |
- |
hypothetical protein CICLE_v100116733mg, partial [Citrus clementina] |
| 9 |
Hb_000108_180 |
0.2130273332 |
- |
- |
PREDICTED: uncharacterized protein LOC105630021 [Jatropha curcas] |
| 10 |
Hb_158092_050 |
0.2159946989 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX10 [Jatropha curcas] |
| 11 |
Hb_001235_120 |
0.2200123876 |
- |
- |
hypothetical protein JCGZ_03395 [Jatropha curcas] |
| 12 |
Hb_000130_350 |
0.2208960484 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 13 |
Hb_000236_230 |
0.222763563 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At4g11680 isoform X2 [Jatropha curcas] |
| 14 |
Hb_166635_010 |
0.2231263237 |
- |
- |
ptm1, putative [Ricinus communis] |
| 15 |
Hb_001541_060 |
0.2242184139 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_116701_010 |
0.2247287177 |
- |
- |
hypothetical protein POPTR_0005s19410g [Populus trichocarpa] |
| 17 |
Hb_019516_070 |
0.229196694 |
- |
- |
PREDICTED: uncharacterized protein LOC105634258 [Jatropha curcas] |
| 18 |
Hb_003177_040 |
0.2298491141 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 19 |
Hb_000251_120 |
0.231522362 |
- |
- |
O-acetyltransferase, putative [Ricinus communis] |
| 20 |
Hb_005695_100 |
0.2352487639 |
- |
- |
PREDICTED: uncharacterized protein LOC105638839 [Jatropha curcas] |