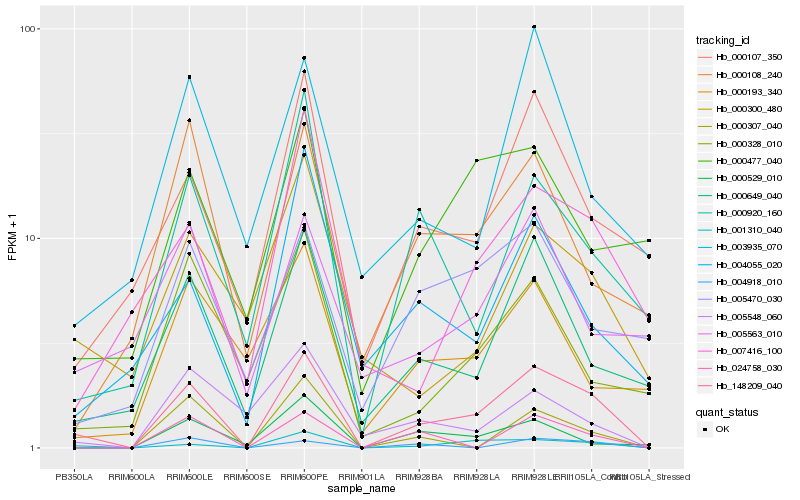
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_148209_040 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001310_040 |
0.185957814 |
transcription factor |
TF Family: TAZ |
transcription cofactor, putative [Ricinus communis] |
| 3 |
Hb_000649_040 |
0.2430348288 |
- |
- |
PREDICTED: dynamin-related protein 1C [Jatropha curcas] |
| 4 |
Hb_000107_350 |
0.2589204929 |
- |
- |
PREDICTED: uncharacterized protein LOC105635576 [Jatropha curcas] |
| 5 |
Hb_024758_030 |
0.2631802826 |
- |
- |
hypothetical protein JCGZ_16050 [Jatropha curcas] |
| 6 |
Hb_000920_160 |
0.2696411247 |
- |
- |
annexin [Manihot esculenta] |
| 7 |
Hb_000307_040 |
0.2749602201 |
- |
- |
glutathione-s-transferase theta, gst, putative [Ricinus communis] |
| 8 |
Hb_000529_010 |
0.2751939006 |
- |
- |
cyclin d, putative [Ricinus communis] |
| 9 |
Hb_000108_240 |
0.2788787883 |
- |
- |
hypothetical protein Csa_1G600150 [Cucumis sativus] |
| 10 |
Hb_004918_010 |
0.2797892608 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 11 |
Hb_007416_100 |
0.2807904748 |
- |
- |
PREDICTED: probable xyloglucan glycosyltransferase 5 [Jatropha curcas] |
| 12 |
Hb_005470_030 |
0.2817932988 |
- |
- |
PREDICTED: probable galactinol--sucrose galactosyltransferase 2 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000328_010 |
0.2826865148 |
- |
- |
PREDICTED: QWRF motif-containing protein 8-like [Jatropha curcas] |
| 14 |
Hb_004055_020 |
0.2829690039 |
- |
- |
PREDICTED: uncharacterized protein LOC105631433 [Jatropha curcas] |
| 15 |
Hb_003935_070 |
0.283133342 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000477_040 |
0.2842453211 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000300_480 |
0.2880532885 |
- |
- |
hypothetical protein RCOM_1321910 [Ricinus communis] |
| 18 |
Hb_005563_010 |
0.2896610887 |
- |
- |
PREDICTED: kinesin-II 85 kDa subunit isoform X1 [Jatropha curcas] |
| 19 |
Hb_005548_060 |
0.2905731643 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000193_340 |
0.2919226322 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 isoform X1 [Jatropha curcas] |