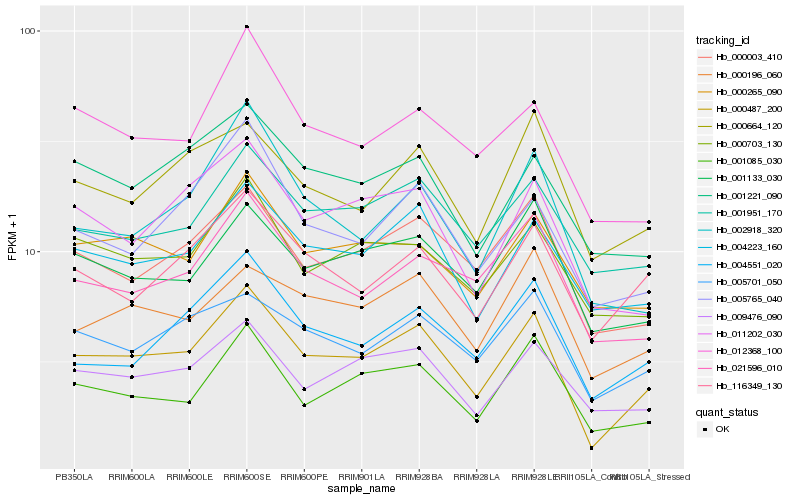
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000487_200 |
0.0 |
- |
- |
- |
| 2 |
Hb_004223_160 |
0.0994253996 |
- |
- |
PREDICTED: 5'-3' exoribonuclease 3 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000003_410 |
0.1000765837 |
- |
- |
PREDICTED: intron-binding protein aquarius [Jatropha curcas] |
| 4 |
Hb_001085_030 |
0.1015448062 |
- |
- |
Tetratricopeptide repeat-like superfamily protein, putative isoform 1 [Theobroma cacao] |
| 5 |
Hb_009476_090 |
0.1054676513 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 1 [Jatropha curcas] |
| 6 |
Hb_011202_030 |
0.1056666338 |
- |
- |
arsenite-resistance protein, putative [Ricinus communis] |
| 7 |
Hb_005701_050 |
0.1070281676 |
- |
- |
PREDICTED: protein LTV1 homolog [Jatropha curcas] |
| 8 |
Hb_004551_020 |
0.107387768 |
- |
- |
PREDICTED: polyadenylation and cleavage factor homolog 4 [Jatropha curcas] |
| 9 |
Hb_001133_030 |
0.1085129618 |
- |
- |
PREDICTED: MAG2-interacting protein 2-like [Jatropha curcas] |
| 10 |
Hb_116349_130 |
0.1090005889 |
transcription factor |
TF Family: Orphans |
PREDICTED: paired amphipathic helix protein Sin3-like 4 isoform X2 [Jatropha curcas] |
| 11 |
Hb_005765_040 |
0.1092923695 |
- |
- |
transcription elongation factor s-II, putative [Ricinus communis] |
| 12 |
Hb_000196_060 |
0.1094365982 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.10-like [Jatropha curcas] |
| 13 |
Hb_000664_120 |
0.109651224 |
- |
- |
PREDICTED: SART-1 family protein DOT2 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000265_090 |
0.1106900506 |
- |
- |
PREDICTED: flowering time control protein FPA [Jatropha curcas] |
| 15 |
Hb_001951_170 |
0.1107158029 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 16 |
Hb_021596_010 |
0.1112912208 |
- |
- |
PREDICTED: uncharacterized protein LOC105647118 [Jatropha curcas] |
| 17 |
Hb_002918_320 |
0.1121029975 |
transcription factor |
TF Family: HB |
PREDICTED: uncharacterized protein LOC105649589 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000703_130 |
0.1121695169 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 [Jatropha curcas] |
| 19 |
Hb_012368_100 |
0.1127314671 |
- |
- |
PREDICTED: uncharacterized protein LOC105637426 [Jatropha curcas] |
| 20 |
Hb_001221_090 |
0.1131379544 |
- |
- |
PREDICTED: uncharacterized protein LOC105640211 isoform X1 [Jatropha curcas] |