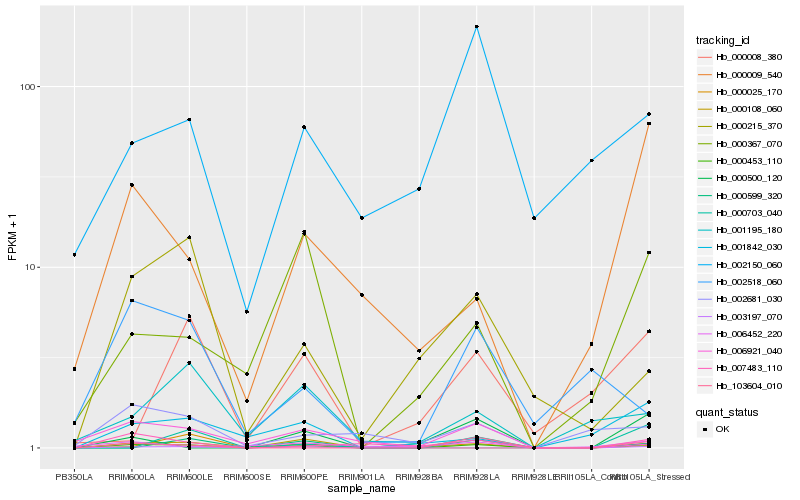
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000453_110 |
0.0 |
- |
- |
PREDICTED: alkaline/neutral invertase CINV2-like isoform X1 [Citrus sinensis] |
| 2 |
Hb_007483_110 |
0.3276942531 |
- |
- |
PREDICTED: beta-xylosidase/alpha-L-arabinofuranosidase 1-like [Populus euphratica] |
| 3 |
Hb_103604_010 |
0.3363578347 |
- |
- |
PREDICTED: uncharacterized protein LOC105795045 [Gossypium raimondii] |
| 4 |
Hb_003197_070 |
0.3504012747 |
- |
- |
PREDICTED: uncharacterized protein LOC104884990 [Beta vulgaris subsp. vulgaris] |
| 5 |
Hb_001842_030 |
0.3577081486 |
- |
- |
PREDICTED: histone deacetylase 9 [Jatropha curcas] |
| 6 |
Hb_000215_370 |
0.3618566097 |
- |
- |
myoinositol oxygenase, putative [Ricinus communis] |
| 7 |
Hb_006921_040 |
0.3677847261 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g25630 [Jatropha curcas] |
| 8 |
Hb_000599_320 |
0.3738552016 |
- |
- |
hypothetical protein JCGZ_17662 [Jatropha curcas] |
| 9 |
Hb_002681_030 |
0.3757005164 |
- |
- |
PREDICTED: fatty-acid-binding protein 1-like [Jatropha curcas] |
| 10 |
Hb_000367_070 |
0.3764207767 |
- |
- |
hypothetical protein JCGZ_11697 [Jatropha curcas] |
| 11 |
Hb_000009_540 |
0.3838023764 |
- |
- |
hypothetical protein JCGZ_12303 [Jatropha curcas] |
| 12 |
Hb_000108_060 |
0.3842247635 |
- |
- |
hypothetical protein POPTR_0010s07820g [Populus trichocarpa] |
| 13 |
Hb_001195_180 |
0.3865793562 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 5 [Jatropha curcas] |
| 14 |
Hb_000025_170 |
0.3870446992 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000500_120 |
0.3908489725 |
- |
- |
PREDICTED: uncharacterized protein LOC105648659 [Jatropha curcas] |
| 16 |
Hb_002518_060 |
0.3997289336 |
- |
- |
PREDICTED: uncharacterized protein LOC105631129 [Jatropha curcas] |
| 17 |
Hb_000703_040 |
0.3999399852 |
- |
- |
hypothetical protein RCOM_1177160 [Ricinus communis] |
| 18 |
Hb_002150_060 |
0.4026116379 |
- |
- |
PREDICTED: arabinogalactan peptide 20-like [Jatropha curcas] |
| 19 |
Hb_006452_220 |
0.4029113769 |
- |
- |
PREDICTED: uncharacterized protein LOC105635043 [Jatropha curcas] |
| 20 |
Hb_000008_380 |
0.4072336564 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |