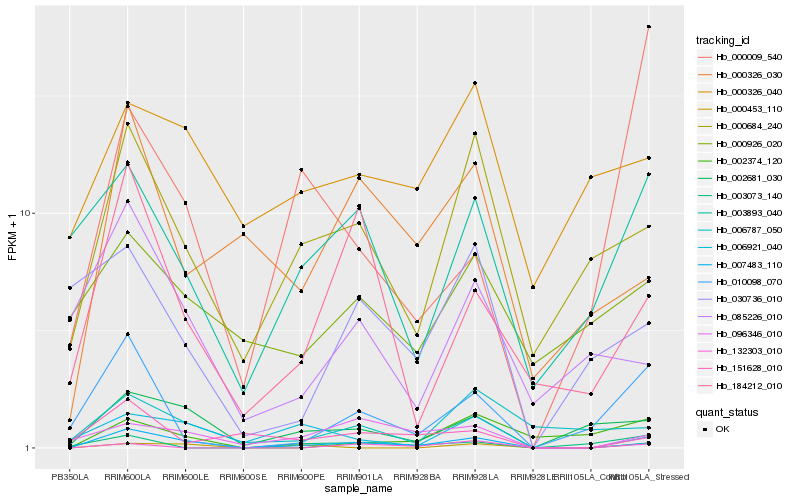
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007483_110 |
0.0 |
- |
- |
PREDICTED: beta-xylosidase/alpha-L-arabinofuranosidase 1-like [Populus euphratica] |
| 2 |
Hb_002681_030 |
0.2815818973 |
- |
- |
PREDICTED: fatty-acid-binding protein 1-like [Jatropha curcas] |
| 3 |
Hb_000684_240 |
0.2950990744 |
- |
- |
hypothetical protein RCOM_0819620 [Ricinus communis] |
| 4 |
Hb_010098_070 |
0.2966704647 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_184212_010 |
0.3202987073 |
- |
- |
PREDICTED: uncharacterized protein LOC105113658 [Populus euphratica] |
| 6 |
Hb_096346_010 |
0.321620266 |
- |
- |
unnamed protein product [Coffea canephora] |
| 7 |
Hb_003893_040 |
0.3225189406 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 8 |
Hb_000453_110 |
0.3276942531 |
- |
- |
PREDICTED: alkaline/neutral invertase CINV2-like isoform X1 [Citrus sinensis] |
| 9 |
Hb_000326_030 |
0.3387624256 |
- |
- |
hypothetical protein PRUPE_ppa013909mg [Prunus persica] |
| 10 |
Hb_151628_010 |
0.3498118535 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X2 [Jatropha curcas] |
| 11 |
Hb_132303_010 |
0.3553466109 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 12 |
Hb_085226_010 |
0.3595442026 |
- |
- |
hypothetical protein POPTR_0002s03930g [Populus trichocarpa] |
| 13 |
Hb_000009_540 |
0.3608440953 |
- |
- |
hypothetical protein JCGZ_12303 [Jatropha curcas] |
| 14 |
Hb_002374_120 |
0.3621664125 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g77405 [Jatropha curcas] |
| 15 |
Hb_006787_050 |
0.364675073 |
- |
- |
hypothetical protein JCGZ_23988 [Jatropha curcas] |
| 16 |
Hb_000326_040 |
0.3663195857 |
- |
- |
hypothetical protein PHAVU_006G187900g [Phaseolus vulgaris] |
| 17 |
Hb_000926_020 |
0.3674375803 |
- |
- |
- |
| 18 |
Hb_003073_140 |
0.3689800135 |
- |
- |
F-box family protein [Theobroma cacao] |
| 19 |
Hb_030736_010 |
0.3696516647 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_006921_040 |
0.3705505384 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g25630 [Jatropha curcas] |