| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
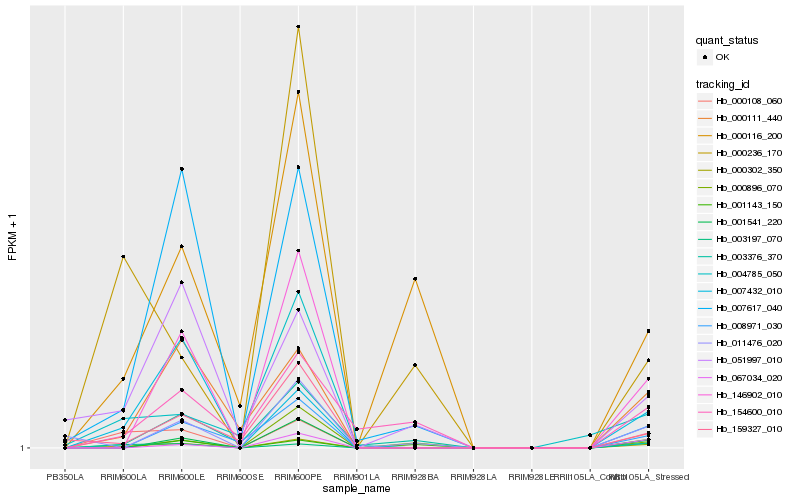
Hb_000302_350 |
0.0 |
- |
- |
polygalacturonase, putative [Ricinus communis] |
| 2 |
Hb_000108_060 |
0.1837809369 |
- |
- |
hypothetical protein POPTR_0010s07820g [Populus trichocarpa] |
| 3 |
Hb_146902_010 |
0.2483191223 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_003197_070 |
0.2537963014 |
- |
- |
PREDICTED: uncharacterized protein LOC104884990 [Beta vulgaris subsp. vulgaris] |
| 5 |
Hb_001541_220 |
0.2557861065 |
transcription factor |
TF Family: ERF |
hypothetical protein RCOM_0793640 [Ricinus communis] |
| 6 |
Hb_000116_200 |
0.2596183788 |
transcription factor |
TF Family: HMG |
hypothetical protein POPTR_0005s10440g [Populus trichocarpa] |
| 7 |
Hb_000111_440 |
0.2614483362 |
- |
- |
- |
| 8 |
Hb_051997_010 |
0.2742808243 |
- |
- |
- |
| 9 |
Hb_154600_010 |
0.2743329526 |
- |
- |
hypothetical protein JCGZ_17786 [Jatropha curcas] |
| 10 |
Hb_007432_010 |
0.2788788515 |
- |
- |
hypothetical protein B456_004G088300 [Gossypium raimondii] |
| 11 |
Hb_067034_020 |
0.2847443042 |
- |
- |
PREDICTED: iridoid synthase [Jatropha curcas] |
| 12 |
Hb_000236_170 |
0.2943201446 |
- |
- |
PREDICTED: Werner Syndrome-like exonuclease [Jatropha curcas] |
| 13 |
Hb_159327_010 |
0.2971734043 |
- |
- |
hypothetical protein JCGZ_23768 [Jatropha curcas] |
| 14 |
Hb_011476_020 |
0.2974865177 |
- |
- |
PREDICTED: glutamate dehydrogenase 2-like [Jatropha curcas] |
| 15 |
Hb_003376_370 |
0.2996387451 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 16 |
Hb_008971_030 |
0.3029559087 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_004785_050 |
0.3034170564 |
- |
- |
PREDICTED: serine decarboxylase [Jatropha curcas] |
| 18 |
Hb_000896_070 |
0.3050997958 |
- |
- |
ammonium transporter, putative [Ricinus communis] |
| 19 |
Hb_007617_040 |
0.3103820522 |
- |
- |
PREDICTED: syntaxin-112-like [Jatropha curcas] |
| 20 |
Hb_001143_150 |
0.3116153239 |
- |
- |
PREDICTED: mitochondrial protein import protein MAS5-like [Populus euphratica] |