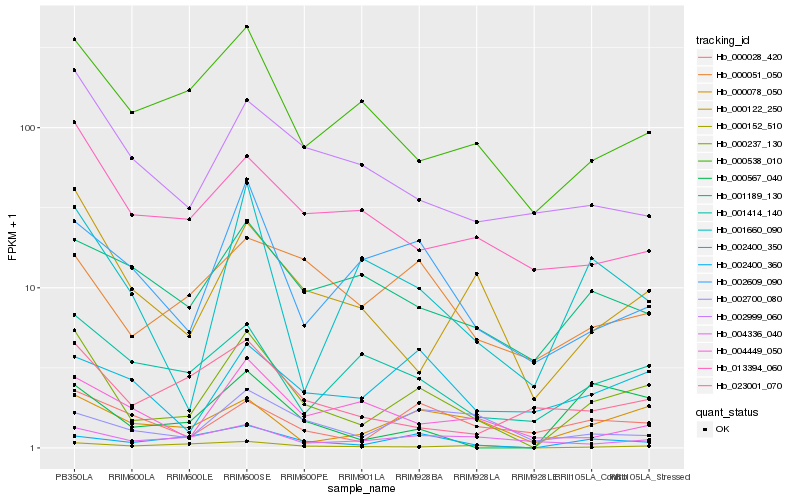
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000237_130 |
0.0 |
- |
- |
- |
| 2 |
Hb_000078_050 |
0.2352185039 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB82 [Jatropha curcas] |
| 3 |
Hb_000122_250 |
0.2365187291 |
transcription factor |
TF Family: ERF |
Dehydration-responsive element-binding protein 2C, putative [Ricinus communis] |
| 4 |
Hb_002400_360 |
0.2370695513 |
- |
- |
heparanase, putative [Ricinus communis] |
| 5 |
Hb_000538_010 |
0.2450983273 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein REVEILLE 1 isoform X2 [Jatropha curcas] |
| 6 |
Hb_002609_090 |
0.2457747781 |
- |
- |
PREDICTED: phosphomethylpyrimidine synthase, chloroplastic isoform X2 [Jatropha curcas] |
| 7 |
Hb_001414_140 |
0.2521808203 |
- |
- |
PREDICTED: probable S-adenosylmethionine-dependent methyltransferase At5g38100 [Populus euphratica] |
| 8 |
Hb_000028_420 |
0.2527261422 |
- |
- |
PREDICTED: uncharacterized protein LOC104886652 [Beta vulgaris subsp. vulgaris] |
| 9 |
Hb_000567_040 |
0.2529211695 |
- |
- |
PREDICTED: uncharacterized protein LOC105649657 [Jatropha curcas] |
| 10 |
Hb_002999_060 |
0.2543508199 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF181 [Jatropha curcas] |
| 11 |
Hb_001660_090 |
0.2552223602 |
- |
- |
PREDICTED: stromal 70 kDa heat shock-related protein, chloroplastic-like [Jatropha curcas] |
| 12 |
Hb_013394_060 |
0.2553138755 |
- |
- |
PREDICTED: IST1-like protein isoform X1 [Jatropha curcas] |
| 13 |
Hb_000152_510 |
0.2564325122 |
- |
- |
kinase, putative [Ricinus communis] |
| 14 |
Hb_000051_050 |
0.2619547379 |
- |
- |
cation efflux protein/ zinc transporter, putative [Ricinus communis] |
| 15 |
Hb_023001_070 |
0.2639086687 |
- |
- |
PREDICTED: dnaJ protein ERDJ3A-like [Eucalyptus grandis] |
| 16 |
Hb_001189_130 |
0.2642019702 |
transcription factor |
TF Family: NAC |
PREDICTED: protein ATAF2-like [Jatropha curcas] |
| 17 |
Hb_004336_040 |
0.2664571761 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 18 |
Hb_002400_350 |
0.2685288502 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 19 |
Hb_004449_050 |
0.2696725032 |
- |
- |
PREDICTED: tyrosine-sulfated glycopeptide receptor 1-like [Jatropha curcas] |
| 20 |
Hb_002700_080 |
0.2737808404 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_24521 [Jatropha curcas] |